Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
O-GlcNAcylation is a highly dynamic, reversible and atypical glycosylation that regulates the activity, biological function, stability, sublocation and interaction of target proteins. O-GlcNAcylation receives and coordinates different signal inputs as an intracellular integrator similar to the nutrient sensor and stress receptor, which target multiple substrates with spatio-temporal analysis specifically to maintain cellular homeostasis and normal physiological functions.
- O-GlcNAc
- skeletal muscle
- metabolism
- insulin resistance
1. Introduction
The proteome is constantly changing and finding harmony with the needs of the organism and its cells, and various PTMs play a unique physiological function in these processes [1][2][3]. Glycosylation, the most extensive and diverse forms of PTMs in eukaryotic cells, contains different types of glycosylation pathways, involves complex metabolic networks and greatly amplifies the proteome by producing the multiple protein forms to instruct a myriad of functions [4][5]. O-GlcNAcylation is a dynamic, reversible and atypical glycosylation [6].
O-GlcNAcylation exists in almost all organisms and is extremely conserved in filamentous fungi, worms, insects, plants and humans [7]. O-GlcNAcylation is also found in all major human organs, even in saliva and urine [8]. O-GlcNAcylation is abundant in the brain, liver, pancreas, skeletal muscle, adipose tissue and other organs and tissues, and plays an essential regulatory role in their physiology and pathology [9][10]. The species distribution of O-GlcNAcylation and its tissues distribution in Homo sapiens are illustrated in Figure 1. O-GlcNAcylation is present in almost all cellular compartments, such as the nucleus, cytoplasm, cytomembrane and mitochondria [11][12]. The distribution characteristics of O-GlcNAcylation also mean that almost all functions of proteins in regulating various cellular processes are covered [13]. O-GlcNAcylated proteins are grouped by protein function as shown in Figure 2. In the past 40 years since O-GlcNAcylation was first discovered, the O-GlcNAcylation of protein has been deeply understood and fruitful results have been obtained. Nearly 5000 human proteins and more than 7000 O-GlcNAcylated sites have been identified in thousands of related research studies [8][14]. O-GlcNAcylation affects the activity, stability, sublocation and biological function of target proteins. The abundance and cycle time scale of O-GlcNAcylation are very similar to that of phosphorylation [15]. Indeed, O-GlcNAcylation has surprisingly extensive crosstalk and forms a yin–yang relationship with phosphorylation, as do acetylation, ubiquitination and other PTMs [16][17]. O-GlcNAcylation receives and integrates metabolic signal pathway inputs from different partners to perceive external environmental disturbance, and ultimately induces adaptive molecular and physiological responses by targeting multiple substrates with the time-space specificity [18][19]. This physiological property makes O-GlcNAcylation extremely sensitive to nutrient availability and environmental changes and to become a nutrient sensor and stress receptor, thus participating in many biological processes [20].
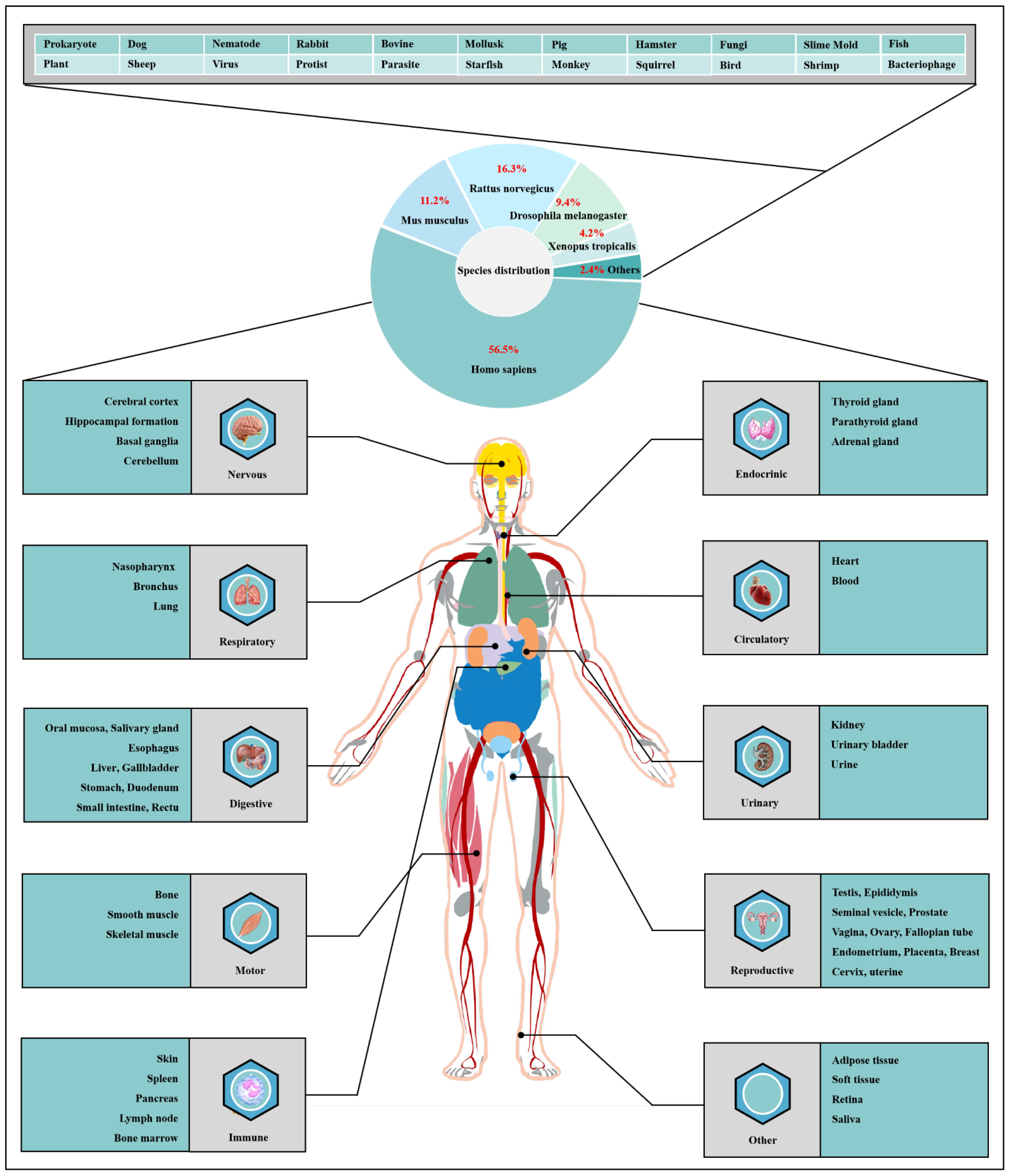
Figure 1. Species distribution of O-GlcNAcylation and its tissues’ distribution in Homo sapiens. The universality and conservation of O-GlcNAcylation is self-evident in filamentous fungi, worms, insects, plants and humans. There are more reports on the O-GlcNAcylation in human, mouse or rat, fruit fly and Caenorhabditis elegans species. However, O-GlcNAcylation has not been identified in yeast, and its similar role may be replaced by the O-mannosylation of nucleocytoplasmic proteins in yeast. O-Glcnacylation was found to occur in almost all major organs of Homo sapiens. This suggests that O-GlcNAcylation is essential for the survival of metazoans and is the root cause of the lethality of OGT and OGA knockout. Tissue distribution analysis emphasizes the strong characteristics of O-GlcNAcylationn in the brain and liver of Homo sapiens. Some organs or tissues with less distribution of O-GlcNAcylation may be limited by their own characteristics of less proteins.
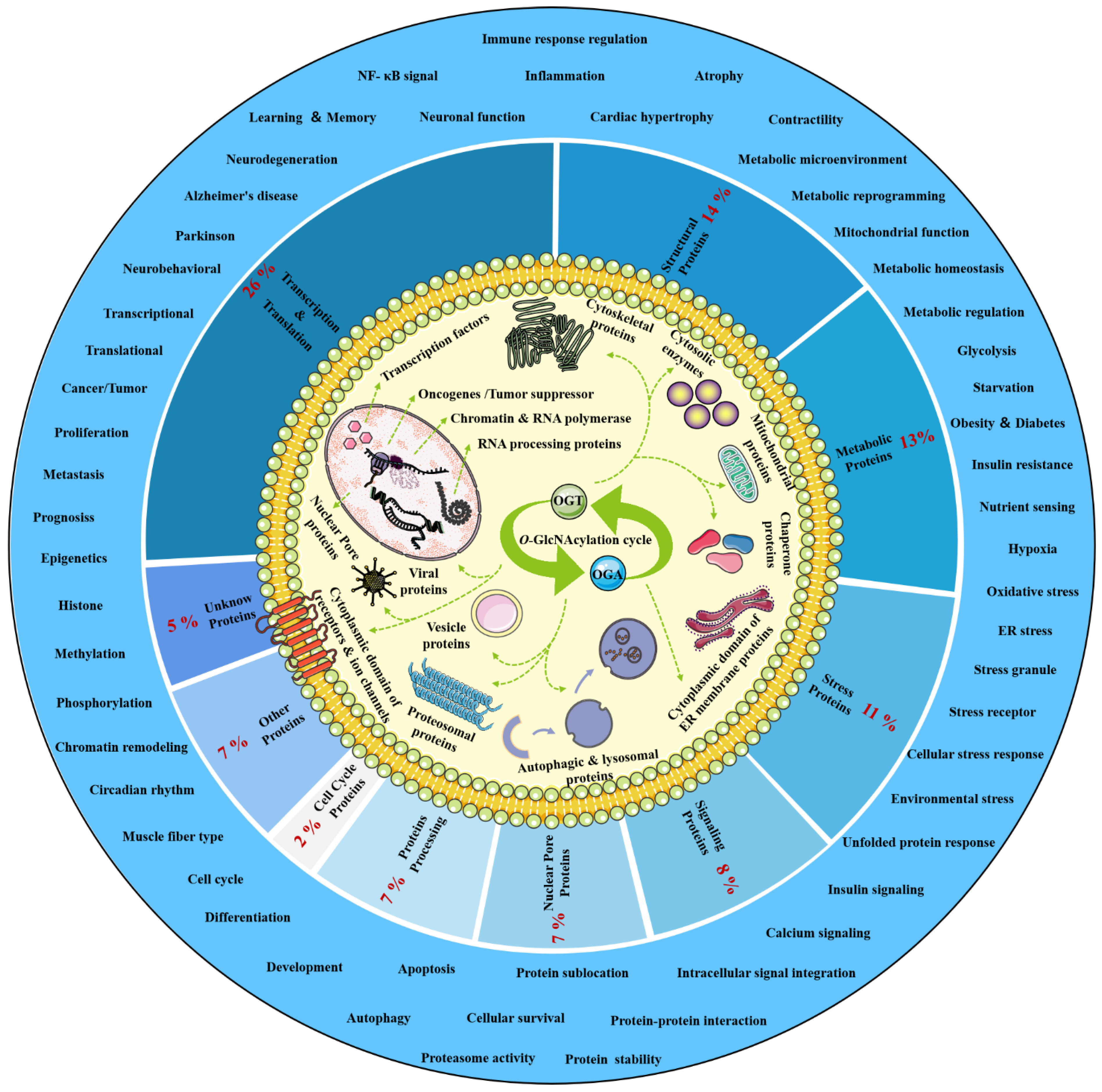
Figure 2. Classification, cellular distribution and physiological process of O-GlcNAcylated proteins. Studies on O-GlcNAcylation have been flourishing since its discovery. To date, a total of 5072 O-GlcNAcylated proteins have been identified by various techniques, and 1803 of these proteins contain 7002 different O-GlcNAcylation sites. However, such a large number may still be only a portion of the abundant dynamically modified proteins within the cellular compartments. The universality and conservation of O-GlcNAcylation is self-evident in filamentous fungi, worms, insects, plants and humans. There are more reports on the O-GlcNAcylation in human, mouse or rat, fruit fly and Caenorhabditis elegans species. However, O-GlcNAcylation has not been identified in yeast, and its similar role may be replaced by the O-mannosylation of nucleocytoplasmic proteins in yeast. These O-GlcNAcylated proteins occur in almost all cellular compartments. O-GlcNAcylated proteins are mainly located in the nuclear and cytoplasmic compartments of all metazoans and their infected viruses. Therefore, O-GlcNAcylation is considered to be one of the most abundant PTMs in the nucleocytoplasmic compartment. Secondly, some mitochondrial proteins are also O-GlcNAcylated. In addition, cytosolic domains of membrane proteins are also O-GlcNAcylated, as well as proteins involved in autophagy and proteosomal degradation of proteins, chaperone proteins, vesicle proteins and numerous cytosolic proteins and enzymes. Meanwhile, the distribution characteristics of O-GlcNAcylation also mean that almost all functions of proteins in regulating various cellular processes are covered. In other words, all functional classes of proteins are affected by O-GlcNAcylation, and these O-GlcNAcylated proteins are distributed according to protein function grouping, as shown above. Some of the largest classes of proteins include those in regulating metabolism, transcription and translation as well as structural proteins. Therefore, O-GlcNAcylation is involved in many cellular processes and pathology, including signal transduction, transcription, translation, chromatin remodeling, protein sublocation and stability, mitochondrial function and cell survival, etc.
2. Dynamic O-GlcNAcylation Cycle and Hexosamine Biosynthesis Pathway
2.1. OGT and OGA Are the Only Antagonistic Enzymes for Precisely Regulating the O-GlcNAcylation Cycle in Space-Time Specificity
OGT has been found to exist in many organisms with high homology. Here, three subtypes of human OGT are taken as an example to demonstrate its gene and protein structure characteristics. OGT has only three isoforms: ncOGT, mimOGT and sOGT [21]. The genomic structure of human OGT is shown in Figure 3A. Ogt is a single gene residing on the chromosome X (Xq13.1) [22]. X inactivation regulates Ogt transcription in female mammals, and Xq13.1 is a region involved in Parkinson’s pathology [23]. Ogt contains 23 exons with alternatively spliced variants [24]. Exon 1 and promoter 1 produce ncOGT. Promoter 2 and the alternative start codon in exon 5 produce mOGT. The alternative start codon in exon 10 generates sOGT. All subtypes of OGT are mainly composed of the amino-terminal TPR domain, the same central linker domain and carboxyl-terminal catalytic domain [25]. All subtypes of OGT are shown in Figure 3B. The three subtypes of OGT contain different numbers of TPRs: 13.5 TPRs in nOGT, 9.5 TPRs in mOGT and 2.5 TPRs in sOGT [26]. Each TPR unit is comprised of 34 amino acid motifs. These TRP units are α-helical and clustered to form a repeating antiparallel helix-turn-helix super spiral [27]. TPRs acts like a “gatekeeper” to identify and interact with the target substrates by contacting their side chains [28]. The mutation of conserved aspartate residues in TPRs has resulted in significant changes in the selectivity, preference and O-GlcNAcylated rate of OGT for target substrates [29]. The influence of TPRs on substrate preference may also be the potential reason for their different cellular localization: mOGT exists in the mitochondria, while ncOGT and sOGT exist in the cytoplasm and nucleus, respectively [30]. In addition, TPRs also has a certain effect on the activity of OGT, and this effect can be exerted only by partial TPRs [31]. TPRs deleting the front 3 or 6 units do not affect the glycosylation of substrates by OGT; however, TPRs deleting the front 9 or 11 units will deactivate OGT [26][32]. OGT belongs to the GT41 gene family of GT-B glycosyltransferase superfamily [33]. Therefore, the carboxyl-terminal catalytic region in OGT has high homology, structural characteristics and catalytic activity of GT-B glycosyltransferases superfamily. Consistent with the structural characteristics of GT-B, OGT contains two similar Rossman folds separated by a deep fissure, which are called CDⅠ and CD Ⅱ, respectively, forming the catalytic core of OGT without the assistance of divalent metal ions [34]. CD Ⅰ mainly consists of a UDP identification pocket and some catalytic groups composed of acidic amino acids that stabilize the pyrophosphate bond through the synergistic action of divalent cations [35]. CD Ⅱ is a lectin-like domain that can be used to recognize and bind the glycoside of UDP-GlcNAc due to its strong affinity for carbohydrates caused by its own abundant trimer structure [36]. However, an Int-D exists in OGT and packs against the carboxyl-terminal catalytic lobe, which is distinct from the structural characteristics of GT-B superfamily [37]. It is preliminarily inferred that Int-D may interact with negatively charged membranes or nucleic acids based on the structural characteristics that contain an unusually large number of surface-exposed basic residues [25]. Meanwhile, PPO is located at the carboxyl terminal of OGT and strongly interacts with PIP3, which promotes the recruitment of OGT to the membrane under insulin induction for catalyzing the dynamic O-GlcNAcylation of the insulin signaling pathway [38]. Targeting special localization of this interaction led to the alteration in the phosphorylation of pivotal insulin signal molecules and weakening of the insulin signal transduction [39]. This suggests the indispensable role of OGT in diabetic pathology. In addition, there is a central flexible linker domain composed of about 120 amino acids [40]. The linker domain seems to exist only in metazoans without high conservatism and its function is unknown, which is the root cause of the difficulty in crystallization of OGT in higher metazoans [41]. The activity and substrate recognition of OGT are also regulated by phosphorylation [42]. OGT is phosphorylated by CHK1 on Ser20, which changes its stabilization and required for cytokinesis [43]. OGT is also phosphorylated by CaMKII on Ser20, which increases its activity [44]. In addition, OGT is also O-GlcNAcylated by itself in Ser3/4 and Thr1045, and their role is unknown [9]. There are multiple acetylation sites in OGT [45]. The presence of these acetylation sites in the catalytic domain suggests that they may modulate OGT activity [14]. The advanced structure of OGT and the sites modified by various PTMs are shown in Figure 3C.
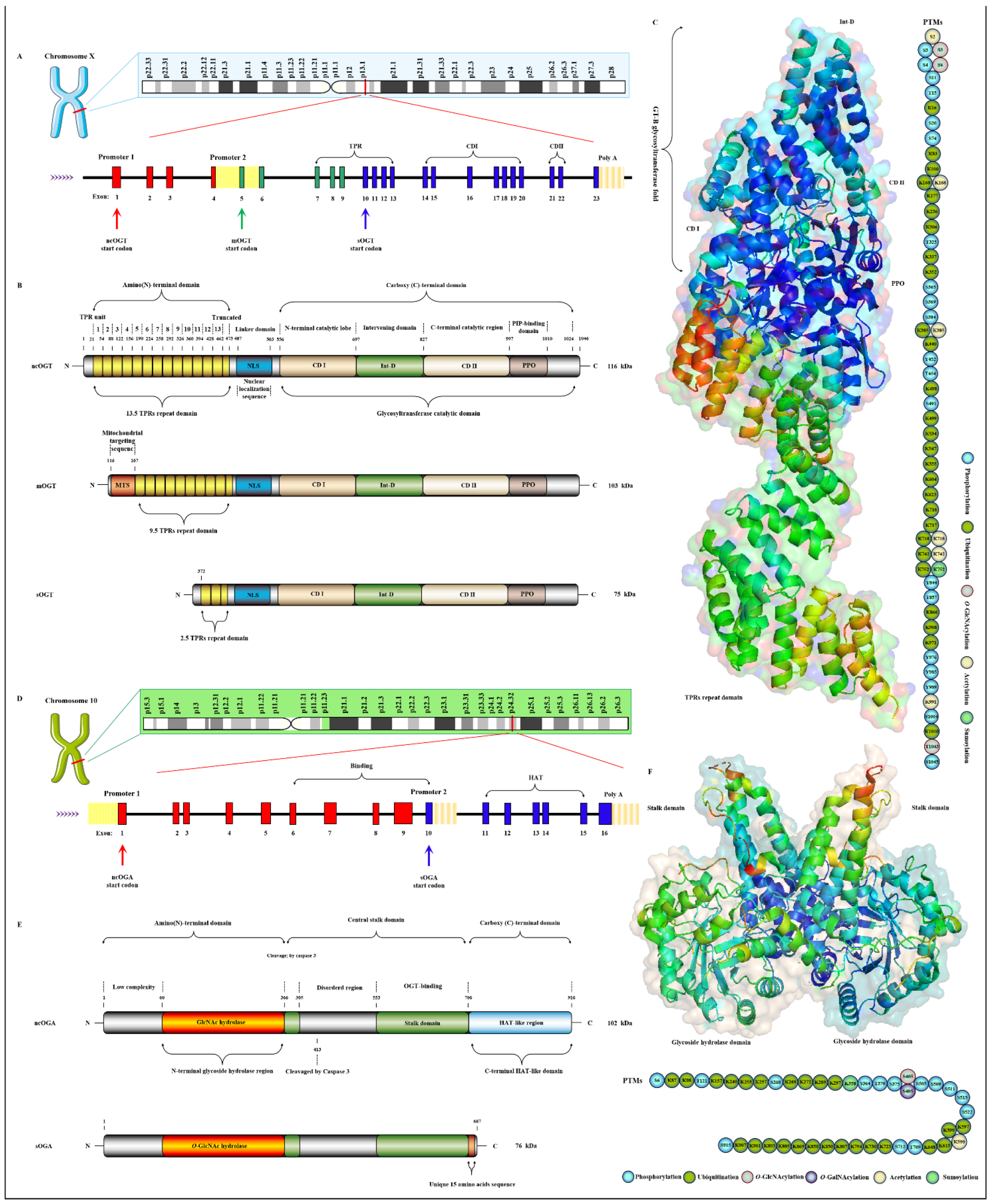
Figure 3. Genomic and proteomic structures of various subtypes of human OGT and OGA. (A) Human Ogt gene mapping and structure. (B) Primary protein structure of three subforms of human OGT. Subtype. (C) Advanced structures of human OGT in 3D and its PTMs. The advanced structure of OGA is displayed in cartoon and surface form with 5M7R in the protein data bank by PyMOL Molecular Graphics System, v2.5.2 (Schrödinger, LLC, New York, NY, USA). The various post-translational modification sites of OGT are present. These predicted modification sites are derived from the PhosphoSitePlus database (https://www.phosphosite.org, (accessed on 10 March 2022). (D) Human Oga gene mapping and structure. (E) Primary protein structure of three subforms of human OGA. Subtype. (F) Advanced structures of human OGA in 3D and its PTMs.
Oga is highly conserved in eukaryotic species, especially in mammals, but absent in prokaryotes and yeast [46]. Oga is mapped to chromosome 10 (10q24.32) as a single gene copy [47]. It is selectively spliced to produce ncOGA and sOGA, which are different at different carboxyl terminals [30]. Gene and protein structures of OGA are shown in Figure 3D,E. Cell fractionation analysis showed that ncOGA was mainly located in the cytoplasm, while the sOGA subtype existed in the nucleus [48]. ncOGA contains the amino-terminal catalytic domain and the central stalk domain and the carboxyl-terminal pseudo-HAT domain linked through two highly disordered (or low complexity) regions [49]. The amino-terminal catalytic domain of OGA is the GlcNAc hydrolysis domain with sequence homology to GH84 [50]. The stalk domain is a hinged region containing multiple alpha helices [51]. It is not conserved between species, which makes it a flexible region that facilitates the folding of the entire protein [52]. Although it has been reported that the HAT-like domain of ncOGA in mice has histone acetyltransferase activity in vitro, it has not been supported by more studies in vivo for lacking the critical residues for the binding of acetyl-coenzyme A [53]. However, the HAT-like domain is evolutionarily conserved, indicating that the pseudo-HAT domain may play an important role in the deglycosylation-associated functions [54]. sOGA lacks the HAT-like domain but contains 15 unique amino acid residues at the carboxyl terminal [55]. Interestingly, it has been reported that sOGA has higher hydrolytic activity in vitro. OGA preferentially removes GlcNAc from some sites, indicating that it has an equal cooperative relationship with OGT in regulating the replacement of O-GlcNAcylation [56]. The active form of OGA appears as homodimer [57]. OGA forms a homodimer in the form of arm to arm, in which the glycoside hydrolase domain of each monomer is covered by the stalk domain of another monomer, thus forming a potential substrate-binding cleft comprising conserved hydrophobic residues [40]. The glycopeptide of the O-GlcNAcylated protein is tightly bound in the substrate-binding cleft through the abundant GlcNAc contacts of the catalytic pocket in OGA, which involves the peptide side chain and the backbone interactions with cleft surface residues [58]. Meanwhile, OGA recognizes the specific characteristics of substrate peptides and hydrolyzes GlcNAc from a wide range of peptide sequences [59]. In addition, some specific residues on OGA contribute to its interaction with different peptide substrates, which means the differential regulation of O-GlcNAcylation on various proteins [60]. OGA is also affected by PTMs such as phosphorylation and O-GlcNAcylation [61]. There are abundant phosphorylation and ubiquitination sites in the domains of glycoside hydrolase and the HAT-like domain [9], but the effect of these modifications at corresponding sites on OGA activity remains to be further determined. The advanced structure of OGA and the sites modified by various PTMs are shown in Figure 3F. The O-GlcNAcylation of OGA at Ser405 is located in the central highly disordered region, suggesting a role in the regulation of OGA-OGT interactions because this is the binding region of OGA-OGT [62]. OGA is also SUMOylated at Lys358 and acetylated at Lys599, respectively [9][63].
2.2. Nutrient Availability Drives Global O-GlcNAcylation through HBP
Extracellular glucose is transported into the intracellular via GLUT-4 [64]. Only 2~3% of the intracellular glucose enters the HBP, while most of the remaining intracellular glucose enters the glycolysis, pentose phosphate pathway (PPP), glycogen synthesis and even polyol pathways, respectively [65]. Therefore, the O-GlcNAcylation cycle is strictly controlled by the flow of glucose through the HBP [66]. Initially, in a study, intracellular glucose was phosphorylated to Glc-6-P by HK, and then Glc-6-P was further isomerized to Fru-6-P by GPI [67]. Subsequently, 3~5% of Fru-6-P was added with an amino group from glutamine to synthesize GlcN-6-P and glutamate by GFAT, while the other 95% of Fru-6-P was used for glycolysis [68]. The enzymatic reaction is the rate-limiting step of HBP, and GFAT is also the key rate-limiting enzyme of HBP [69]. The activity of GFAT is still regulated by multiple pathways [70]. Firstly, the activity of GFAT is regulated by substrate availability, which is positively activated by the concentration of glucose and glutamine, and the negative feedback is inhibited by the concentration of UDP-GlcNAc and GlcN-6-P [71]. The activity of GFAT is also closely related to some PTMs. The Ser243 of GFAT is phosphorylated and its activity is reduced by AMPK, mTORC2 and CaMKII, and a similar effect is also caused by 2-Deoxy-D-glucose [72][73]. PKA also promotes the phosphorylation of GFAT at Ser205/235 [74]. Succinylation of GFAT at Lys529, acetylation of GFAT at Lys114, 547, 650 and multiple ubiquitination Lys sites of GFAT are predicted by PhosphoSitePlus® v6.6.0.2 (https://www.phosphosite.org, accessed on 10 March 2022). Meanwhile, it has been reported that specificity protein 1, activating transcription factor 4 and X-box-binding protein 1 regulate GFAT at the transcriptional level [75][76]. Glutamine is necessary for this enzymatic reaction, but this restriction can be bypassed by glucosamine as an extended supplement [77]. Therefore, incubating cells with glucosamine or high concentration glucose or glutamine can bypass the rate-limiting step catalyzed by GFAT, thereby increasing global O-GlcNAcylation. GNA converts GlcNAc-6-P using acetyl-CoA [78]. Then, GlcNAc-6-P is catalytically translocated to GlcNAc-1-P by AGM [79]. It is worth noting that the only difference of HBP in prokaryotes is that GlcN-6-P is isomerized to GlcN-1-P and then GlcN-1-P is acetylated to form GlcNAc-1-P [80]. The HBP process in eukaryotes is as shown above. Finally, UTP is then utilized by UAP to convert GlcNAc-1-P into UDP-GlcNAc and release iPPi [81]. The HBP process involves the participation of glucose, glutamine, uridine, acetyl-CoA and ATP [82]. Therefore, UDP-GlcNAc, as the end-product of HBP, integrates the metabolisms of carbohydrates, amino acids, fats and nucleotides [83]. UDP-GlcNAc is a unique donor of O-GlcNAcylation, which provides GlcNAc, which is necessary and irreplaceable for O-GlcNAcylation [84]. GlcNAc provided by UDP-GlcNAc is used and transferred by OGT to the oxygen atom of the hydroxyl group of serine or threonine residues of the target protein [85]. On the contrary, the GlcNAc moiety is removed from O-GlcNAcylated proteins by OGA [86]. These hydrolyzed GlcNAc or other free GlcNAc obtained by lysosomal or nutrient degradation are converted to GlcNAc-6-P through N-Acetylglucosamine kinase (NAGK) and then used again for the synthesis of UDP-GlcNAc [87]. Therefore, GlcNAc can also bypass the rate-limiting step of HBP and GFAT, which is also effective for salvage pathways such as glucosamine and glutamine [88]. In addition, UDP-GlcNAc is also used as a substrate for the synthesis of proteoglycans, hyaluronic acid, glycolipids, GPI anchor, N-glycosylation and other O-glycosylation [89]. The activated UDP-GlcNAc is utilized by concentration-sensitive enzymes in the nucleus, cytoplasm and membrane to glycosylate the substrate or generate glucose conjugates [89]. UDP-GlcNAc is actively transported by nucleotide sugar transporters to cellular organelles, such as the ER and Golgi apparatus [90]. The differences in UDP-GlcNA permeability and relative cell volume of these organelles complicate the estimation of the cytoplasmic and nuclear concentrations of UDP-GlcNAc [91]. The relative abundance of O-GlcNAcylation is roughly negatively correlated with the more complex glycans [92]. These characteristics make UDP-GlcNAc and its derivatives extremely sensitive to the variations in cellular nutrients, so that the dynamic O-GlcNAcylaion can be used as a reporter of the functional status of multiple pathways and regarded as a metabolic sensor [20]. Meanwhile, the mutual conversion and complex relationship of the intermediate products in the HBP, polyol pathway, PPP, glycogen, glycolysis and TCA cycle intermediates greatly enlarge the nutritional sensitivity of O-GlcNAcylation [93] and also suggest the potential mechanism of O-GlcNAcylaion’s negative feedback regulation of these glucose metabolism branches. Indeed, O-GlcNAcylation is involved in multiple modes of metabolic regulation. Almost all the enzymes involved in glycolysis were identified to have been modified by O-GlcNAcylation [94]. The O-GlcNAcylated enzymes exist in every step of glycolysis, including GLUT4, HK, GPI, PFK, FBA, GAPDH, PGK, PDM, ENO, PK and PDC [95][96][97][98]. Glycogen synthesis is also regulated by O-GlcNAcylated GSK3β, and PPP activity is affected by O-GlcNAcylated G6PD [99][100]. In addition, increased HBP flux and O-GlcNAcylation also promotes fatty acid oxidation in the heart and adipose tissue [101]. The O-GlcNAylation of several transcription factors, such as PGC1α, FoxO3, NF-κB and CREB, also indirectly participates in transcriptional regulation of metabolism [102][103][104]. Although only briefly shown in Figure 4, it is worth noting that almost all enzymes in the TCA cycle are also modified by O-GlcNAcylation, such as AH, IDH, KGD, SL, SDH, MDH and the several subunits of respiratory chain complexes [105][106]. CS and FH may be potentially O-GlcNAcylatied, but there is still a lack of supporting evidence [94].
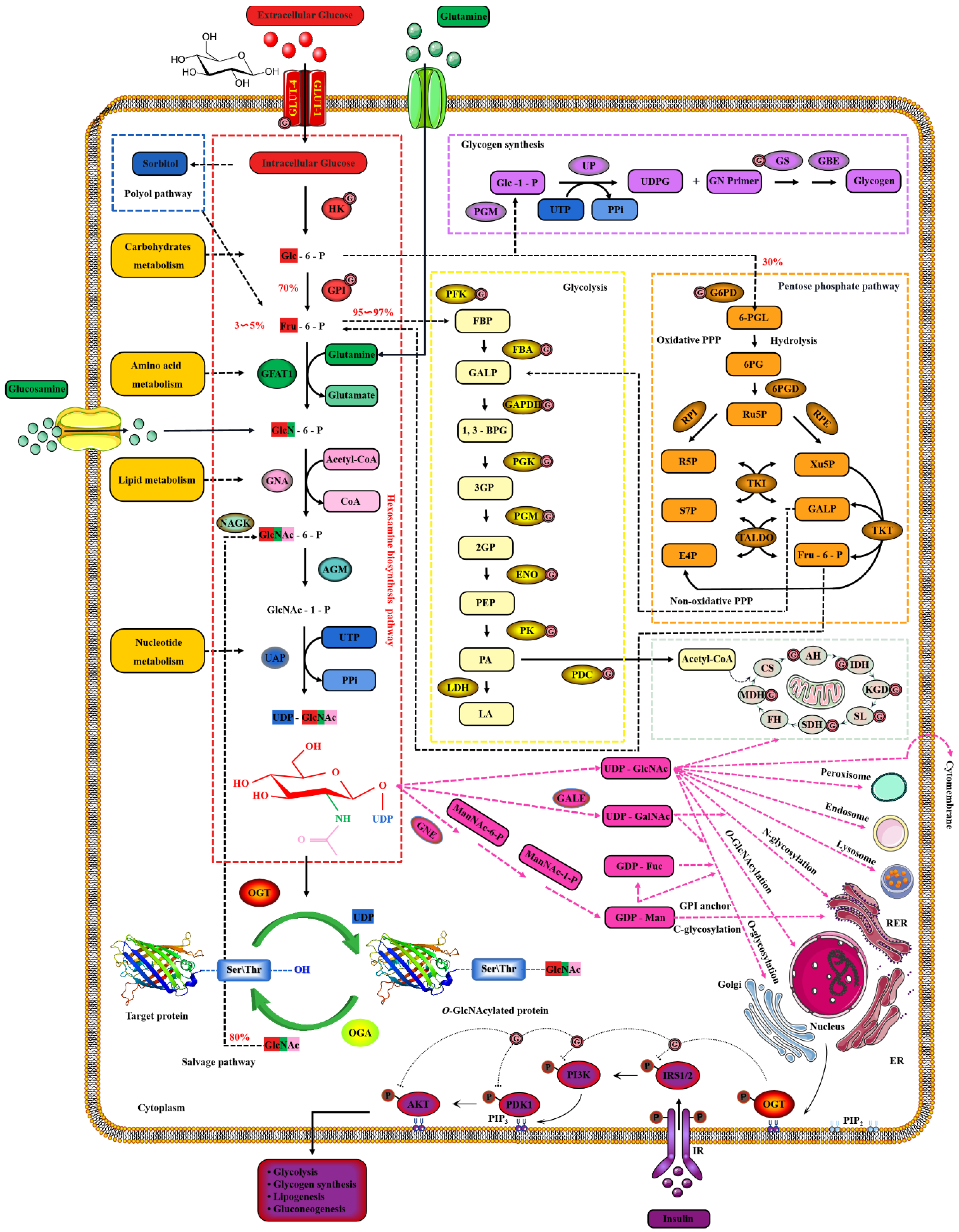
Figure 4. Nutrient availability drives the dynamic cycle of protein O-GlcNAcylation via the hexosamine biosynthesis pathway. The GlcNAc provided by UDP-GlcNAc is necessary and unique for O-GlcNAcylation, and the only source of UDP-GlcNAc is HBP. Changes in nutritional availability, such as carbohydrates, lipids, amino acids, nucleotides and ATP, fluctuate HBP flux. Therefore, UDP-GlcNAc and HBP link carbohydrate metabolism, lipid metabolism, amino acid metabolism and nucleotide metabolism. These physiological characteristics make OGT extremely sensitive to nutrient fluctuations.
3. O-GlcNAcylation, Energy Metabolism and Insulin Sensitivity in Skeletal Muscle
Skeletal muscle is a repository of nutrients, enabling it to serve as the consumer and producer of energy during exercise, stress and starvation [107]. The energy requirements of skeletal muscle are enormous, and its energy expenditure increases 300 times from the base state to the full contraction state [108][109]. This directly affects glucose homeostasis in skeletal muscle, where CaMKII or GLUT4 is activated to increase glucose uptake [110]. In fact, skeletal muscle processes more than 80% of insulin to stimulate glucose uptake and is considered as one of the most critical insulin-sensitive tissues [111]. The metabolic flexibility in skeletal muscle ensures an adequate supply of energy for its work [112]. O-GlcNAcylation as a cellular nutrient sensor plays a key role in glucose metabolism during this physiological process [113]. In addition, skeletal muscle is the main target organ of insulin, and nutrient-driven O-GlcNAcylation is a key regulator of insulin signaling in skeletal muscle [114]. O-GlcNAcylation is considered to be critical in the dysregulation of the insulin signaling cascade and the molecular mechanism of insulin resistance [115].
4. O-GlcNAcylation Is an Emerging Mediator of Contractile and Structural Properties in Skeletal Muscle
4.1. O-GlcNAcylation Is an Essential Regulator of Contractile Properties in Skeletal Muscle
To date, many O-GlcNAcylated contractile proteins and contractile-related regulatory proteins in sarcomere have been identified [116]. These key contractile proteins include actin, myosin, MLC and MHC, tropomyosin and troponin, etc. [117][118][119]. In view of this, the physiological role of O-GlcNAcylation on skeletal muscle contractile activity has been concerned. Many data emphasize that O-GlcNAcylation mediates calcium activation properties to regulate the contractile activity of skeletal muscle. Increased O-GlcNAcylation of MHC, α-tropomyosin and α-sarcomeric actin in myocardium of diabetic mice resulted in the decrease of sarcomere calcium sensitivity [120]. Reversible reductions in calcium affinity and sensitivity of muscular fibers occur when exposed to GlcNAc, and O-GlcNAcylation of some critical contractile proteins increased, such as MHC, MLC and actin [121]. Moreover, similar results appeared in skinned fibers and cardiac trabeculae. Phosphorylation of Tn I at the Ser23 and Ser24 by PKA improves the calcium sensitivity of cardiomyocytes and alter of myofilament properties [122]. Interestingly, Tn I, Tn T and Tn C of the troponin complex are O-GlcNAcylated in skeletal muscle and myocardial tissue [123]. The O-GlcNAcylation levels of Tn I in fiber cells exposed to GlcNAc or OGA inhibitors increased, and the calcium activation properties reduced [124]. However, this treatment did not affect the phosphorylation levels of Tn I at Ser23/24 [125]. One of the possible mechanisms is that the GlcNAc part destroys the protein–protein interaction [119].
O-GlcNAcyation of MLC2 in myocardial tissue occurred at the site of Ser15. It is worth noting that the only phosphorylation site overlaps with the only O-GlcNAcyation site on MLC2. This suggests that the close potential interaction between O-GlcNAcyation and phosphorylation of MLC2 is in the calcium activation properties of sarcomere. This interaction has been proved to be mutually exclusive, and the dynamics of this interaction vary according to the pattern of skeletal muscle activity [126]. The precise regulation of phosphorylation and O-GlcNAcyation on MLC2 involves a multienzyme cluster. This multienzyme cluster contains MLCK/MYPT2/PP1 and OGT/OGA, which are involved in the phosphorylation and O-GlcNAcyation of MLC2 [127]. This multienzyme cluster is preferentially located at the Z disk of the sarcomere and responds to the physiological signals of skeletal muscle to strengthen the interaction between the enzymes contained in itself [128]. For example, the partial recombination of this multienzyme clusters in skeletal muscle dysfunction, such as increased co-localization between MLCK and OGA. The location of OGT and OGA in the diabetic heart is redistributed in the sarcomere [129]. This is due to enhanced OGA activity and increased the interaction with α-actin, tropomyosin and MLC1, while OGT was the opposite. This results in the removal of abnormal O-GlcNAcylation to restore myofilaments to Ca2+ response [130]. Nevertheless, the exact effect and mechanism of O-GlcNAcylation on MLC2 need to be further confirmed. One credible assumption is that O-GlcNAcylation causes steric hindration between MLC2 and MHC due to its stokes radius being many times larger than phosphate [15].
In general, O-GlcNAcylation is considered as a new mechanism to regulate the contractile properties of skeletal muscle by modifying critical contractile proteins to regulate the interaction between other proteins or with itself, and phosphorylation to mediate calcium activation. In addition, the regulatory effect of O-GlcNAcylation on the contractile activity of skeletal muscle is also related to its effect on sarcomere structure [131], which isdiscussed in the next section.
4.2. O-GlcNAcylation Is an Emerging Maintainer of the Structural Properties in Skeletal Muscle
O-GlcNAcylation also regulates the polymerization of desmin, just as it regulates the polymerization of tubulin and cytokeratin filaments 8/18 [132][133][134]. α/β-crystallin serve as molecular chaperones to facilitate the localization, aggregation, and assembly of desmin [135]. O-GlcNAcylation of α/β-crystallin at Thr170 and Thr162 regulates its localization and its interaction with desmin, respectively [136][137]. The multiple O-GlcNAcylation sites of mouse titin are located on the kelch-12 domain, and the absence of titin leads to the change of muscle structure and the decrease of muscle performance [138][139]. These O-GlcNAcylation sites are located in the key regions of sarcomere assembly and myosin polymerization and its interaction with MyBP-C and MHC [140][141]. Interestingly, O-GlcNAcylation sites of MHC have also been found. These sites are adjacent to its domain of polymerization and interaction with myosin and titin [142]. O-GlcNAcylation of MHC is located at Ser1708, and it is further considered to be involved in Laing early-onset distal myopathy due to its proximity to mutant Leu1706 residue. Meanwhile, O-GlcNAcylation sites in the PxxP domain of BAG3 and the plakin domain repeat B5 of plectin have been identified, which are related to the interaction with SH3-containing protein and intermediate filament proteins, respectively [138]. This evidence suggests that O-GlcNAcylation occurs in specific domains of certain structural proteins to regulate interactions other proteins for maintaining sarcomere structure and function. This view was confirmed by a recent study that O-GlcNAcylated milton binds to FHL2 to anchor mitochondria to F-actin [143][144]. The main targets and pathways of O-GlcNAcylation in skeletal muscle physiology are shown in Figure 5.
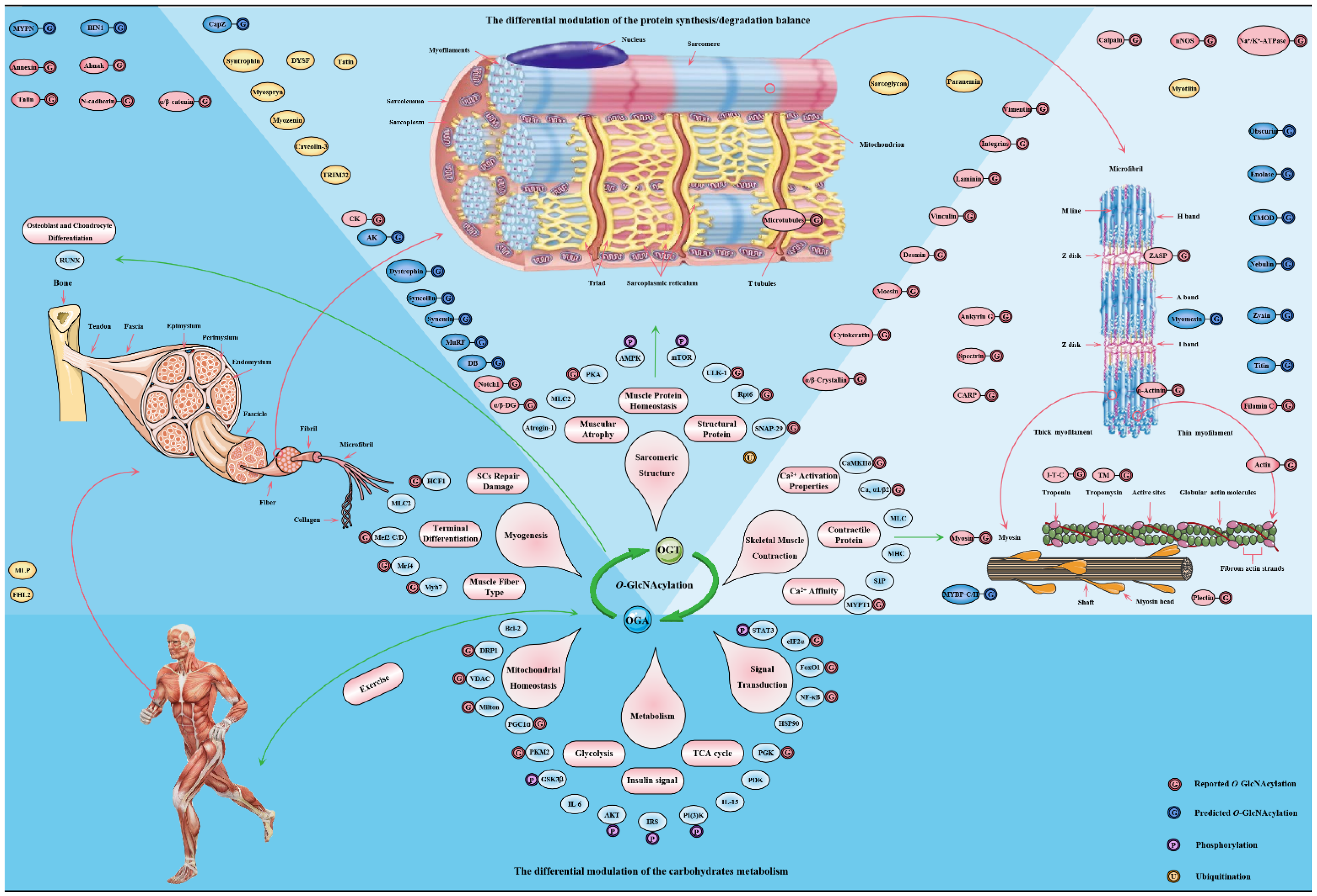
Figure 5. The major targets and pathways shown to be altered by O-GlcNAcylation in in the skeletal muscle physiology. Thousands of O-GlcNAcylated proteins have been identified in skeletal muscle cells. These O-GlcNAcylated proteins are classified into contractile proteins, sarcolemma proteins, structural proteins and cytoskeletal proteins, as well as mitochondrial proteins, metabolic enzymes, transcription factors and signal proteins. Therefore, the effects of O-GlcNAcylation on various physiological processes of skeletal muscle may be realized from the following four aspects: (1) its regulation of carbohydrate metabolism with sensing nutritional availability; (2) its maintenance of structural protein synthesis/degradation balance; (3) its improvement of sarcomere contractile activity by modulating the calcium activation properties; (4) its promotion of adaptation and protection under exercise and certain adverse circumstances.
This entry is adapted from the peer-reviewed paper 10.3390/cells11111789
References
- Conibear, A.C. Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 2020, 4, 674–695.
- Liu, J.; Qian, C.; Cao, X. Post-Translational Modification Control of Innate Immunity. Immunity 2016, 45, 15–30.
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214.
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749.
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366.
- Yang, X.; Qian, A.K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465.
- Itkonen, H.M.; Loda, M.; Mills, I.G. O-GlcNAc Transferase—An Auxiliary Factor or a Full-blown Oncogene? Mol Cancer Res. 2021, 19, 555–564.
- Wulff-Fuentes, E.; Berendt, R.R.; Massman, L.; Danner, L.; Malard, F.; Vora, J.; Kahsay, R.; Olivier-Van Stichelen, S. The human O-GlcNAcome database and meta-analysis. Sci. Data 2021, 8, 25.
- Chatham, J.C.; Zhang, J.; Wende, A.R. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 2021, 101, 427–493.
- Morino, K.; Maegawa, H. Role of O-linked N-acetylglucosamine in the homeostasis of metabolic organs, and its potential links with diabetes and its complications. J. Diabetes Investig. 2021, 12, 130–136.
- Eustice, M.; Bond, M.R.; Hanover, J.A. O-GlcNAc cycling and the regulation of nucleocytoplasmic dynamics. Biochem. Soc. Trans. 2017, 45, 427–436.
- Sakaidani, Y.; Nomura, T.; Matsuura, A.; Ito, M.; Suzuki, E.; Murakami, K.; Nadano, D.; Matsuda, T.; Furukawa, K.; Okajima, T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2011, 2, 583.
- Li, Y.; Xie, M.; Men, L.; Du, J. O-GlcNAcylation in immunity and inflammation: An intricate system (Review). Int J. Mol. Med. 2019, 44, 363–374.
- Malard, F.; Wulff-Fuentes, E.; Berendt, R.R.; Didier, G.; Olivier-Van Stichelen, S. Automatization and self-maintenance of the O-GlcNAcome catalog: A smart scientific database. Database 2021, 2021, baab039.
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858.
- Yuan, A.; Tang, X.; Li, J. Centrosomes: Til O-GlcNAc Do Us Apart. Front. Endocrinol. 2020, 11, 621888.
- Cui, Z.; Scruggs, S.B.; Gilda, J.E.; Ping, P.; Gomes, A.V. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J. Mol. Cell. Cardiol. 2014, 71, 32–42.
- Ong, Q.; Han, W.; Yang, X. O-GlcNAc as an Integrator of Signaling Pathways. Front. Endocrinol. 2018, 9, 599.
- Zhu, Y.; Hart, G.W. Targeting O-GlcNAcylation to develop novel therapeutics. Mol. Aspects Med. 2021, 79, 100885.
- Liu, Y.; Yao, R.Z.; Lian, S.; Liu, P.; Hu, Y.J.; Shi, H.Z.; Lv, H.M.; Yang, Y.Y.; Xu, B.; Li, S.Z. O-GlcNAcylation: The “stress and nutrition receptor” in cell stress response. Cell Stress Chaperones 2021, 26, 297–309.
- Kim, E.J.; Abramowitz, L.K.; Bond, M.R.; Love, D.C.; Kang, D.W.; Leucke, H.F.; Kang, D.W.; Ahn, J.S.; Hanover, J.A. Versatile O-GlcNAc transferase assay for high-throughput identification of enzyme variants, substrates, and inhibitors. Bioconjug. Chem. 2014, 25, 1025–1030.
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739.
- Olivier-Van Stichelen, S.; Hanover, J.A. X-inactivation normalizes O-GlcNAc transferase levels and generates an O-GlcNAc-depleted Barr body. Front Genet. 2014, 5, 256.
- Nolte, D.; Müller, U. Human O-GlcNAc transferase (OGT): Genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm. Genome 2002, 13, 62–64.
- Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469, 564–567.
- Iyer, S.P.; Hart, G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003, 278, 24608–24616.
- Ramirez, D.H.; Yang, B.; D’Souza, A.K.; Shen, D.; Woo, C.M. Truncation of the TPR domain of OGT alters substrate and glycosite selection. Anal. Bioanal. Chem. 2021, 413, 7385–7399.
- Levine, Z.G.; Walker, S. The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells? Annu. Rev. Biochem. 2016, 85, 631–657.
- Joiner, C.M.; Levine, Z.G.; Aonbangkhen, C.; Woo, C.M.; Walker, S. Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. J. Am. Chem. Soc. 2019, 141, 12974–12978.
- Gloster, T.M.; Vocadlo, D.J. Mechanism, Structure, and Inhibition of O-GlcNAc Processing Enzymes. Curr. Signal Transduct. Ther. 2010, 5, 74–91.
- Clarke, A.J.; Hurtado-Guerrero, R.; Pathak, S.; Schüttelkopf, A.W.; Borodkin, V.; Shepherd, S.M.; Ibrahim, A.F.; van Aalten, D.M. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008, 27, 2780–2788.
- Rafie, K.; Raimi, O.; Ferenbach, A.T.; Borodkin, V.S.; Kapuria, V.; van Aalten, D.M.F. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017, 7.
- Pathak, S.; Alonso, J.; Schimpl, M.; Rafie, K.; Blair, D.E.; Borodkin, V.S.; Albarbarawi, O.; van Aalten, D.M.F. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 2015, 22, 744–750.
- Shi, J.; Ruijtenbeek, R.; Pieters, R.J. Demystifying O-GlcNAcylation: Hints from peptide substrates. Glycobiology 2018, 28, 814–824.
- She, N.; Zhao, Y.; Hao, J.; Xie, S.; Wang, C. Uridine diphosphate release mechanism in O-N-acetylglucosamine (O-GlcNAc) transferase catalysis. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 609–622.
- Lazarus, B.D.; Roos, M.D.; Hanover, J.A. Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J. Biol. Chem. 2005, 280, 35537–35544.
- Fan, Q.; Moen, A.; Anonsen, J.H.; Bindesbøll, C.; Sæther, T.; Carlson, C.R.; Grønning-Wang, L.M. O-GlcNAc site-mapping of liver X receptor-α and O-GlcNAc transferase. Biochem. Biophys. Res. Commun. 2018, 499, 354–360.
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969.
- Moore, M.; Avula, N.; Jo, S.; Beetch, M.; Alejandro, E.U. Disruption of O-Linked N-Acetylglucosamine Signaling in Placenta Induces Insulin Sensitivity in Female Offspring. Int. J. Mol. Sci. 2021, 22, 6918.
- Gorelik, A.; van Aalten, D.M.F. Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem. Biol. 2020, 1, 98–109.
- Hurtado-Guerrero, R.; Dorfmueller, H.C.; van Aalten, D.M. Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 2008, 18, 551–557.
- Hardivillé, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213.
- Li, Z.; Li, X.; Nai, S.; Geng, Q.; Liao, J.; Xu, X.; Li, J. Checkpoint kinase 1-induced phosphorylation of O-linked β-N-acetylglucosamine transferase regulates the intermediate filament network during cytokinesis. J. Biol. Chem. 2017, 292, 19548–19555.
- Ruan, H.B.; Ma, Y.; Torres, S.; Zhang, B.; Feriod, C.; Heck, R.M.; Qian, K.; Fu, M.; Li, X.; Nathanson, M.H.; et al. Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 2017, 31, 1655–1665.
- Seo, H.G.; Kim, H.B.; Kang, M.J.; Ryum, J.H.; Yi, E.C.; Cho, J.W. Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep. 2016, 6, 34614.
- Alonso, J.; Schimpl, M.; van Aalten, D.M. O-GlcNAcase: Promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J. Biol. Chem. 2014, 289, 34433–34439.
- Kim, E.J. Chemical arsenal for the study of O-GlcNAc. Molecules 2011, 16, 1987–2022.
- Elbatrawy, A.A.; Kim, E.J.; Nam, G. Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nat. Commun. 2017, 8, 666.
- Elbatrawy, A.A.; Kim, E.J.; Nam, G. O-GlcNAcase: Emerging Mechanism, Substrate Recognition and Small-Molecule Inhibitors. ChemMedChem 2020, 15, 1244–1257.
- Elsen, N.L.; Patel, S.B.; Ford, R.E.; Hall, D.L.; Hess, F.; Kandula, H.; Kornienko, M.; Reid, J.; Selnick, H.; Shipman, J.M.; et al. Insights into activity and inhibition from the crystal structure of human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 613–615.
- Macauley, M.S.; Vocadlo, D.J. Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 2010, 1800, 107–121.
- Kohler, J.J. Carb cutting works better with a partner. Nat. Struct. Mol. Biol. 2017, 24, 433–435.
- Rao, F.V.; Schüttelkopf, A.W.; Dorfmueller, H.C.; Ferenbach, A.T.; Navratilova, I.; van Aalten, D.M. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 2013, 3, 130021.
- He, Y.; Roth, C.; Turkenburg, J.P.; Davies, G.J. Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase. Acta Crystallogr. D Biol. Crystallogr. 2014, 70 Pt 1, 186–195.
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880.
- Martin, J.C.; Fadda, E.; Ito, K.; Woods, R.J. Defining the structural origin of the substrate sequence independence of O-GlcNAcase using a combination of molecular docking and dynamics simulation. Glycobiology 2014, 24, 85–96.
- de Alencar, N.A.; Sousa, P.R.; Silva, J.R.; Lameira, J.; Alves, C.N.; Martí, S.; Moliner, V. Computational analysis of human OGA structure in complex with PUGNAc and NAG-thiazoline derivatives. J. Chem. Inf. Model 2012, 52, 2775–2783.
- Roth, C.; Chan, S.; Offen, W.A.; Hemsworth, G.R.; Willems, L.I.; King, D.T.; Varghese, V.; Britton, R.; Vocadlo, D.J.; Davies, G.J. Structural and functional insight into human O-GlcNAcase. Nat. Chem. Biol. 2017, 13, 610–612.
- Schimpl, M.; Schüttelkopf, A.W.; Borodkin, V.S.; van Aalten, D.M. Human OGA binds substrates in a conserved peptide recognition groove. Biochem. J. 2010, 432, 1–7.
- Kim, E.J. In Vitro Biochemical Assays for O-GlcNAc-Processing Enzymes. Chembiochem 2017, 18, 1462–1472.
- Wu, D.; Cai, Y.; Jin, J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 2017, 8, 713–723.
- Balsollier, C.; Pieters, R.J.; Anderluh, M. Overview of the Assays to Probe O-Linked β-N-Acetylglucosamine Transferase Binding and Activity. Molecules 2021, 26, 1037.
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431.
- Flores-Opazo, M.; McGee, S.L.; Hargreaves, M. Exercise and GLUT4. Exerc. Sport Sci. Rev. 2020, 48, 110–118.
- Richter, E.A.; Hargreaves, M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol. Rev. 2013, 93, 993–1017.
- Hanover, J.A.; Krause, M.W.; Love, D.C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 2010, 1800, 80–95.
- Adeva-Andany, M.M.; Pérez-Felpete, N.; Fernández-Fernández, C.; Donapetry-García, C.; Pazos-García, C. Liver glucose metabolism in humans. Biosci Rep. 2016, 36, e00416.
- Filhoulaud, G.; Guillemain, G.; Scharfmann, R. The hexosamine biosynthesis pathway is essential for pancreatic beta cell development. J. Biol. Chem. 2009, 284, 24583–24594.
- Love, D.C.; Hanover, J.A. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE 2005, 2005, re13.
- Zibrova, D.; Vandermoere, F.; Göransson, O.; Peggie, M.; Mariño, K.V.; Knierim, A.; Spengler, K.; Weigert, C.; Viollet, B.; Morrice, N.A.; et al. GFAT1 phosphorylation by AMPK promotes VEGF-induced angiogenesis. Biochem. J. 2017, 474, 983–1001.
- Ruegenberg, S.; Horn, M.; Pichlo, C.; Allmeroth, K.; Baumann, U.; Denzel, M.S. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat. Commun. 2020, 11, 687.
- Leto, D.; Saltiel, A.R. Regulation of glucose transport by insulin: Traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 2012, 13, 383–396.
- Eguchi, S.; Oshiro, N.; Miyamoto, T.; Yoshino, K.; Okamoto, S.; Ono, T.; Kikkawa, U.; Yonezawa, K. AMP-activated protein kinase phosphorylates glutamine: Fructose-6-phosphate amidotransferase 1 at Ser243 to modulate its enzymatic activity. Genes Cells 2009, 14, 179–189.
- Ruegenberg, S.; Mayr, F.; Atanassov, I.; Baumann, U.; Denzel, M.S. Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1. Nat. Commun. 2021, 12, 2176.
- Al-Mukh, H.; Baudoin, L.; Bouaboud, A.; Sanchez-Salgado, J.L.; Maraqa, N.; Khair, M.; Pagesy, P.; Bismuth, G.; Niedergang, F.; Issad, T. Lipopolysaccharide Induces GFAT2 Expression to Promote O-Linked β-N-Acetylglucosaminylation and Attenuate Inflammation in Macrophages. J. Immunol. 2020, 205, 2499–2510.
- Horn, M.; Denzel, S.I.; Srinivasan, B.; Allmeroth, K.; Schiffer, I.; Karthikaisamy, V.; Miethe, S.; Breuer, P.; Antebi, A.; Denzel, M.S. Hexosamine Pathway Activation Improves Protein Homeostasis through the Integrated Stress Response. iScience 2020, 23, 100887.
- Denzel, M.S.; Antebi, A. Hexosamine pathway and (ER) protein quality control. Curr. Opin. Cell Biol. 2015, 33, 14–18.
- Zachara, N.E.; Hart, G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004, 1673, 13–28.
- Biwi, J.; Biot, C.; Guerardel, Y.; Vercoutter-Edouart, A.S.; Lefebvre, T. The Many Ways by Which O-GlcNAcylation May Orchestrate the Diversity of Complex Glycosylations. Molecules 2018, 23, 2858.
- Lam, C.; Low, J.Y.; Tran, P.T.; Wang, H. The hexosamine biosynthetic pathway and cancer: Current knowledge and future therapeutic strategies. Cancer Lett. 2021, 503, 11–18.
- Olivier-Van Stichelen, S.; Hanover, J.A. You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 339–345.
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231.
- Coussement, P.; Bauwens, D.; Peters, G.; Maertens, J.; De Mey, M. Mapping and refactoring pathway control through metabolic and protein engineering: The hexosamine biosynthesis pathway. Biotechnol. Adv. 2020, 40, 107512.
- Vincenz, L.; Hartl, F.U. Sugarcoating ER Stress. Cell 2014, 156, 1125–1127.
- Gurel, Z.; Sheibani, N. O-Linked β-N-acetylglucosamine (O-GlcNAc) modification: A new pathway to decode pathogenesis of diabetic retinopathy. Clin. Sci. 2018, 132, 185–198.
- Saha, A.; Bello, D.; Fernández-Tejada, A. Advances in chemical probing of protein O-GlcNAc glycosylation: Structural role and molecular mechanisms. Chem. Soc. Rev. 2021, 50, 10451–10485.
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019, 17, 52.
- Chatham, J.C.; Nöt, L.G.; Fülöp, N.; Marchase, R.B. Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock 2008, 29, 431–440.
- Vasconcelos-Dos-Santos, A.; Oliveira, I.A.; Lucena, M.C.; Mantuano, N.R.; Whelan, S.A.; Dias, W.B.; Todeschini, A.R. Biosynthetic Machinery Involved in Aberrant Glycosylation: Promising Targets for Developing of Drugs Against Cancer. Front. Oncol. 2015, 5, 138.
- Denzel, M.S.; Storm, N.J.; Gutschmidt, A.; Baddi, R.; Hinze, Y.; Jarosch, E.; Sommer, T.; Hoppe, T.; Antebi, A. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell 2014, 156, 1167–1178.
- Groves, J.A.; Lee, A.; Yildirir, G.; Zachara, N.E. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 2013, 18, 535–558.
- Bolanle, I.O.; Palmer, T.M. Targeting Protein O-GlcNAcylation, a Link between Type 2 Diabetes Mellitus and Inflammatory Disease. Cells 2022, 11, 705.
- Ma, Z.; Vosseller, K. O-GlcNAc in cancer biology. Amino Acids 2013, 45, 719–733.
- Bacigalupa, Z.A.; Bhadiadra, C.H.; Reginato, M.J. O-GlcNAcylation: Key regulator of glycolytic pathways. J. Bioenerg. Biomembr. 2018, 50, 189–198.
- Lambert, M.; Bastide, B.; Cieniewski-Bernard, C. Involvement of O-GlcNAcylation in the Skeletal Muscle Physiology and Physiopathology: Focus on Muscle Metabolism. Front. Endocrinol. 2018, 9, 578.
- Nie, H.; Ju, H.; Fan, J.; Shi, X.; Cheng, Y.; Cang, X.; Zheng, Z.; Duan, X.; Yi, W. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 2020, 11, 36.
- Singh, J.P.; Qian, K.; Lee, J.S.; Zhou, J.; Han, X.; Zhang, B.; Ong, Q.; Ni, W.; Jiang, M.; Ruan, H.B.; et al. O-GlcNAcase targets pyruvate kinase M2 to regulate tumor growth. Oncogene 2020, 39, 560–573.
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A., 3rd; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980.
- Maury, J.J.; Ng, D.; Bi, X.; Bardor, M.; Choo, A.B. Multiple reaction monitoring mass spectrometry for the discovery and quantification of O-GlcNAc-modified proteins. Anal. Chem. 2014, 86, 395–402.
- Ou, W.; Liang, Y.; Qin, Y.; Wu, W.; Xie, M.; Zhang, Y.; Zhang, Y.; Ji, L.; Yu, H.; Li, T. Hypoxic acclimation improves cardiac redox homeostasis and protects heart against ischemia-reperfusion injury through upregulation of O-GlcNAcylation. Redox Biol. 2021, 43, 101994.
- Baldini, S.F.; Lefebvre, T. O-GlcNAcylation and the Metabolic Shift in High-Proliferating Cells: All the Evidence Suggests that Sugars Dictate the Flux of Lipid Biogenesis in Tumor Processes. Front. Oncol. 2016, 6, 6.
- Benhamed, F.; Filhoulaud, G.; Caron, S.; Lefebvre, P.; Staels, B.; Postic, C. O-GlcNAcylation Links ChREBP and FXR to Glucose-Sensing. Front. Endocrinol. 2014, 5, 230.
- Lim, K.; Yoon, B.H.; Ha, C.H. O-Linked N-acetylglucosaminylation of Sp1 interferes with Sp1 activation of glycolytic genes. Biochem. Biophys. Res. Commun. 2015, 468, 349–353.
- Kuo, M.; Zilberfarb, V.; Gangneux, N.; Christeff, N.; Issad, T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008, 582, 829–834.
- Reggiori, F.; Gabius, H.J.; Aureli, M.; Römer, W.; Sonnino, S.; Eskelinen, E.L. Glycans in autophagy, endocytosis and lysosomal functions. Glycoconj J. 2021, 38, 625–647.
- Cao, W.; Cao, J.; Huang, J.; Yao, J.; Yan, G.; Xu, H.; Yang, P. Discovery and confirmation of O-GlcNAcylated proteins in rat liver mitochondria by combination of mass spectrometry and immunological methods. PLoS ONE 2013, 8, e76399.
- Deshmukh, A.S. Insulin-stimulated glucose uptake in healthy and insulin-resistant skeletal muscle. Horm. Mol. Biol. Clin. Investig. 2016, 26, 13–24.
- Jørgensen, S.B.; Richter, E.A.; Wojtaszewski, J.F. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J. Physiol. 2006, 574, 17–31.
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828.
- Ojuka, E.O.; Goyaram, V.; Smith, J.A. The role of CaMKII in regulating GLUT4 expression in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E322–E331.
- Merz, K.E.; Thurmond, D.C. Role of Skeletal Muscle in Insulin Resistance and Glucose Uptake. Compr. Physiol. 2020, 10, 785–809.
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184.
- Hortemo, K.H.; Lunde, P.K.; Anonsen, J.H.; Kvaløy, H.; Munkvik, M.; Rehn, T.A.; Sjaastad, I.; Lunde, I.G.; Aronsen, J.M.; Sejersted, O.M. Exercise training increases protein O-GlcNAcylation in rat skeletal muscle. Physiol. Rep. 2016, 4, e12896.
- Shi, H.; Munk, A.; Nielsen, T.S.; Daughtry, M.R.; Larsson, L.; Li, S.; Høyer, K.F.; Geisler, H.W.; Sulek, K.; Kjøbsted, R.; et al. Skeletal muscle O-GlcNAc transferase is important for muscle energy homeostasis and whole-body insulin sensitivity. Mol. Metab. 2018, 11, 160–177.
- Teo, C.F.; Wollaston-Hayden, E.E.; Wells, L. Hexosamine flux, the O-GlcNAc modification, and the development of insulin resistance in adipocytes. Mol. Cell. Endocrinol. 2010, 318, 44–53.
- Lambert, M.; Claeyssen, C.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation as a regulator of the functional and structural properties of the sarcomere in skeletal muscle: An update review. Acta Physiol. 2020, 228, e13301.
- Stevens, L.; Bastide, B.; Hedou, J.; Cieniewski-Bernard, C.; Montel, V.; Cochon, L.; Dupont, E.; Mounier, Y. Potential regulation of human muscle plasticity by MLC2 post-translational modifications during bed rest and countermeasures. Arch. Biochem. Biophys. 2013, 540, 125–132.
- Akimoto, Y.; Yan, K.; Miura, Y.; Tsumoto, H.; Toda, T.; Fukutomi, T.; Sugahara, D.; Kudo, A.; Arai, T.; Chiba, Y.; et al. O-GlcNAcylation and phosphorylation of β-actin Ser(199) in diabetic nephropathy. Am. J. Physiol. Renal. Physiol. 2019, 317, F1359–F1374.
- Ramirez-Correa, G.A.; Jin, W.; Wang, Z.; Zhong, X.; Gao, W.D.; Dias, W.B.; Vecoli, C.; Hart, G.W.; Murphy, A.M. O-linked GlcNAc modification of cardiac myofilament proteins: A novel regulator of myocardial contractile function. Circ. Res. 2008, 103, 1354–1358.
- Ramirez-Correa, G.A.; Ma, J.; Slawson, C.; Zeidan, Q.; Lugo-Fagundo, N.S.; Xu, M.; Shen, X.; Gao, W.D.; Caceres, V.; Chakir, K.; et al. Removal of Abnormal Myofilament O-GlcNAcylation Restores Ca2+ Sensitivity in Diabetic Cardiac Muscle. Diabetes 2015, 64, 3573–3587.
- Hedou, J.; Cieniewski-Bernard, C.; Leroy, Y.; Michalski, J.C.; Mounier, Y.; Bastide, B. O-linked N-acetylglucosaminylation is involved in the Ca2+ activation properties of rat skeletal muscle. J. Biol. Chem. 2007, 282, 10360–10369.
- Salhi, H.E.; Hassel, N.C.; Siddiqui, J.K.; Brundage, E.A.; Ziolo, M.T.; Janssen, P.M.; Davis, J.P.; Biesiadecki, B.J. Myofilament Calcium Sensitivity: Mechanistic Insight into TnI Ser-23/24 and Ser-150 Phosphorylation Integration. Front. Physiol. 2016, 7, 567.
- Metzger, J.M.; Westfall, M.V. Covalent and noncovalent modification of thin filament action: The essential role of troponin in cardiac muscle regulation. Circ. Res. 2004, 94, 146–158.
- Cieniewski-Bernard, C.; Montel, V.; Berthoin, S.; Bastide, B. Increasing O-GlcNAcylation level on organ culture of soleus modulates the calcium activation parameters of muscle fibers. PLoS ONE 2012, 7, e48218.
- Bayliss, C.R.; Jacques, A.M.; Leung, M.C.; Ward, D.G.; Redwood, C.S.; Gallon, C.E.; Copeland, O.; McKenna, W.J.; Dos Remedios, C.; Marston, S.B.; et al. Myofibrillar Ca2+ sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc. Res. 2013, 97, 500–508.
- Hortemo, K.H.; Aronsen, J.M.; Lunde, I.G.; Sjaastad, I.; Lunde, P.K.; Sejersted, O.M. Exhausting treadmill running causes dephosphorylation of sMLC2 and reduced level of myofilament MLCK2 in slow twitch rat soleus muscle. Physiol. Rep. 2015, 3, e12285.
- Cieniewski-Bernard, C.; Dupont, E.; Richard, E.; Bastide, B. Phospho-GlcNAc modulation of slow MLC2 during soleus atrophy through a multienzymatic and sarcomeric complex. Pflugers Arch. 2014, 466, 2139–2151.
- Cai, L.X.; Tanada, Y.; Bello, G.D.; Fleming, J.C.; Alkassis, F.F.; Ladd, T.; Golde, T.; Koh, J.; Chen, S.; Kasahara, H. Cardiac MLC2 kinase is localized to the Z-disc and interacts with α-actinin2. Sci. Rep. 2019, 9, 12580.
- Wang, L.; Geist, J.; Grogan, A.; Hu, L.R.; Kontrogianni-Konstantopoulos, A. Thick Filament Protein Network, Functions, and Disease Association. Compr. Physiol. 2018, 8, 631–709.
- Wende, A.R. Unsticking the Broken Diabetic Heart: O-GlcNAcylation and Calcium Sensitivity. Diabetes 2015, 64, 3339–3341.
- Rassier, D.E. Sarcomere mechanics in striated muscles: From molecules to sarcomeres to cells. Am. J. Physiol. Cell Physiol. 2017, 313, C134–C145.
- Srikanth, B.; Vaidya, M.M.; Kalraiya, R.D. O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J. Biol. Chem. 2010, 285, 34062–34071.
- Ise, H.; Kobayashi, S.; Goto, M.; Sato, T.; Kawakubo, M.; Takahashi, M.; Ikeda, U.; Akaike, T. Vimentin and desmin possess GlcNAc-binding lectin-like properties on cell surfaces. Glycobiology 2010, 20, 843–864.
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids 2011, 40, 809–818.
- Lin, B.L.; Song, T.; Sadayappan, S. Myofilaments: Movers and Rulers of the Sarcomere. Compr. Physiol. 2017, 7, 675–692.
- Ma, J.; Wang, W.H.; Li, Z.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. O-GlcNAc Site Mapping by Using a Combination of Chemoenzymatic Labeling, Copper-Free Click Chemistry, Reductive Cleavage, and Electron-Transfer Dissociation Mass Spectrometry. Anal. Chem. 2019, 91, 2620–2625.
- Krishnamoorthy, V.; Donofrio, A.J.; Martin, J.L. O-GlcNAcylation of αB-crystallin regulates its stress-induced translocation and cytoprotection. Mol. Cell. Biochem. 2013, 379, 59–68.
- Deracinois, B.; Camoin, L.; Lambert, M.; Boyer, J.B.; Dupont, E.; Bastide, B.; Cieniewski-Bernard, C. O-GlcNAcylation site mapping by (azide-alkyne) click chemistry and mass spectrometry following intensive fractionation of skeletal muscle cells proteins. J. Proteom. 2018, 186, 83–97.
- Sziklai, D.; Sallai, J.; Papp, Z.; Kellermayer, D.; Mártonfalvi, Z.; Pires, R.H.; Kellermayer, M.S.Z. Nanosurgical Manipulation of Titin and Its M-Complex. Nanomaterials 2022, 12, 178.
- Kontrogianni-Konstantopoulos, A.; Ackermann, M.A.; Bowman, A.L.; Yap, S.V.; Bloch, R.J. Muscle giants: Molecular scaffolds in sarcomerogenesis. Physiol. Rev. 2009, 89, 1217–1267.
- Li, Y.; Lang, P.; Linke, W.A. Titin stiffness modifies the force-generating region of muscle sarcomeres. Sci. Rep. 2016, 6, 24492.
- Hédou, J.; Bastide, B.; Page, A.; Michalski, J.C.; Morelle, W. Mapping of O-linked beta-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics 2009, 9, 2139–2148.
- Basu, H.; Pekkurnaz, G.; Falk, J.; Wei, W.; Chin, M.; Steen, J.; Schwarz, T.L. FHL2 anchors mitochondria to actin and adapts mitochondrial dynamics to glucose supply. J. Cell Biol. 2021, 220, e201912077.
- Canault, M.; Tellier, E.; Bonardo, B.; Mas, E.; Aumailley, M.; Juhan-Vague, I.; Nalbone, G.; Peiretti, F. FHL2 interacts with both ADAM-17 and the cytoskeleton and regulates ADAM-17 localization and activity. J. Cell. Physiol. 2006, 208, 363–372.
This entry is offline, you can click here to edit this entry!
