Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Transposable elements (TEs) are mobile DNA sequences that can jump from one genomic locus to another and that have colonized the genomes of all living organisms. While TE mobilization is an important source of genomic innovations that greatly contribute to the host species evolution, it is also a major threat to genome integrity that can lead to pathologies.
- transposable elements
- endogenous retroviruses
- horizontal transfer
1. Introduction
Transposable elements (TEs) are DNA sequences that can move and multiply within the genome by transposition. They were discovered by Barbara McClintock in the maize genome in 1950 [1]. Since then, TEs have been found in all living organisms in which they have been searched for. The TE community is still debating whether there are organisms without TE sequences in their genome. Their amplification within genomes leads to the formation of families of repeated sequences that can be present, for some of them, in thousands of copies spread in all chromosomes. They are currently classified in two categories [2]: (i) class I elements, also referred to as retrotransposons, that transpose through a copy-and-paste mechanism, and (ii) class II elements, referred to as DNA transposons, that transpose through a cut-and-paste mechanism (excision and re-insertion at a new locus). TE transposition is a major source of genetic instability, especially through chromosome breakages and insertions that result in mutations, ectopic recombination, and genomic rearrangements. To preserve genome integrity, TE mobilization is strictly controlled by several elaborate defense mechanisms, such as silencing strategies based on KRAB zinc-finger proteins, small RNAs, DNA methylation, and chromatin modifications [3][4][5][6][7]. Moreover, most transposon sequences accumulate mutations that do not allow them to produce the proteins required for their transposition and that may ultimately lead to elimination of all active copies of that mobile element in a population. The combined actions of silencing, mutations, and elimination should result in the complete elimination of TEs from the genome. Yet, TEs represent a large part of the genome in all organisms (prokaryotes, unicellular and multicellular eukaryotes), ranging from 3% in yeast to 85% in maize [8]. Indeed, it is assumed that TE movement and accumulation are an important source of genomic and epigenomic variations that strongly influence the species evolution and adaptation to changing environments [9][10][11][12]. However, it remains unclear how TEs persist in the genome, reach such high proportions, and expand in all living species, while transposition is strictly controlled.
2. Genome Invasion by Transposable Elements: Strategies for Effective Spreading
2.1. Horizontal Transfer: TE Propagation between Species
A major step in understanding how TEs might persist over time was the discovery that some TEs can colonize “naive” genomes through horizontal transfer (HT). HT is the transmission of genetic material between closely or distantly related organisms in the absence of reproduction. These events permit the acquisition of exogenous genetic material and, therefore, are responsible for the appearance of genetic novelties. The first evidence of HT involving TEs (Horizontal Transposon Transfer, HTT) in eukaryotes was the HT and subsequent invasion by the P-element, a DNA transposon, between two fruit fly species (i.e., from Drosophila willistoni to Drosophila melanogaster). P-elements rapidly spread through natural populations of D. melanogaster between 1950 and 1980, and all flies collected in the wild after 1980 have P-elements, unlike laboratory strains derived from flies collected before 1950 [13][14][15][16]. The P-element in the D. melanogaster genome differs by only one nucleotide from that in the D. willistoni genome. This demonstrated that the P-element found in D. willistoni was transferred to D. melanogaster some time before 1950. Currently, the number of fully sequenced genomes is sufficiently high to reveal that HTT is a widespread phenomenon in metazoans. For instance, more than 500 putative HTT events have been described between Drosophila species [17]. In insects, HTT is not an anecdotic event because up to 24% of all nucleotides of insect genomes might come from HTTs [17]. The same authors showed that DNA transposons transfer horizontally more frequently than retrotransposons. These findings indicate that HTT is a fundamental mechanism implicated in eukaryotic genome evolution. It allows TEs to bypass the host silencing machinery by introduction into naive species that have not yet adapted to silence new TEs.
TE mobility and replication characteristics may facilitate the invasion and integration into the host genome. However, the precise mechanisms by which TEs can be shuttled between organisms and the nature of the potential vectors remain speculative (Figure 1 panel 1). It has been suggested that host–parasite interactions favor HTT. For instance, the P-elements could have been transmitted from D. willistoni to D. melanogaster thanks to the mite Proctolaelaps regalis [18]. During feeding by piercing and sucking the fly eggs and larvae, this mite might transmit genetic material (e.g., DNA transposons) from one fly to another. Insects, such as wasps or the hemipteran Rhodnius prolixus, that feed on the blood of mammals, birds, and reptiles might be involved in HT as vectors (reviewed in [19]). Bacteria and viruses also might be interesting vectors of HTT between species. Indeed, their capacity to transfer DNA and recombine with the host genome might allow them to transport various TE sequences from host to host. Gilbert et al. analyzed 21 genomes of a baculovirus population and demonstrated that a substantial number of TEs from the infected host can transpose into the baculovirus genome [20]. The discovery of a retroposon sequence (Short Interspersed Nucleotide Element, SINE) and its flanking regions coming from the genome of a West African snake (Echis ocellatus) in the genome of the taterapox virus (TATV) is another piece of evidence that viruses are frequently used as vectors for HTT [21]. Wolbachia is an intracellular parasitic bacterium that infects mainly arthropod species and also some nematodes. This bacterium transfers vertically and horizontally between species and can also transit from cell to cell and infect the host germ cells. Interestingly, many gene transfer events have been detected between Drosophila and Wolbachia, suggesting that this bacterium is a good candidate vector for HTT between arthropods [22][23]. As a final example, it has been proposed that nematodes may be both great vectors for HTT but also serve as TE reservoirs [24]. Nematodes are ubiquitous organisms and the geographical proximity with many different species increased their chance to participate either as a donor or as a recipient in HTT events. In line with that, many horizontal transfer events involving TEs have been reported to occur between nematodes and unrelated organisms. Future phylogenetic studies will probably reveal many other HTT events involving many different mechanisms and vectors. Combined with geographical and ecological data, these findings will help to unravel the complex and dynamic network of gene transfer and HTT that break down taxon boundaries, and to determine the contribution of ancient HTT events to the evolution of different organisms.
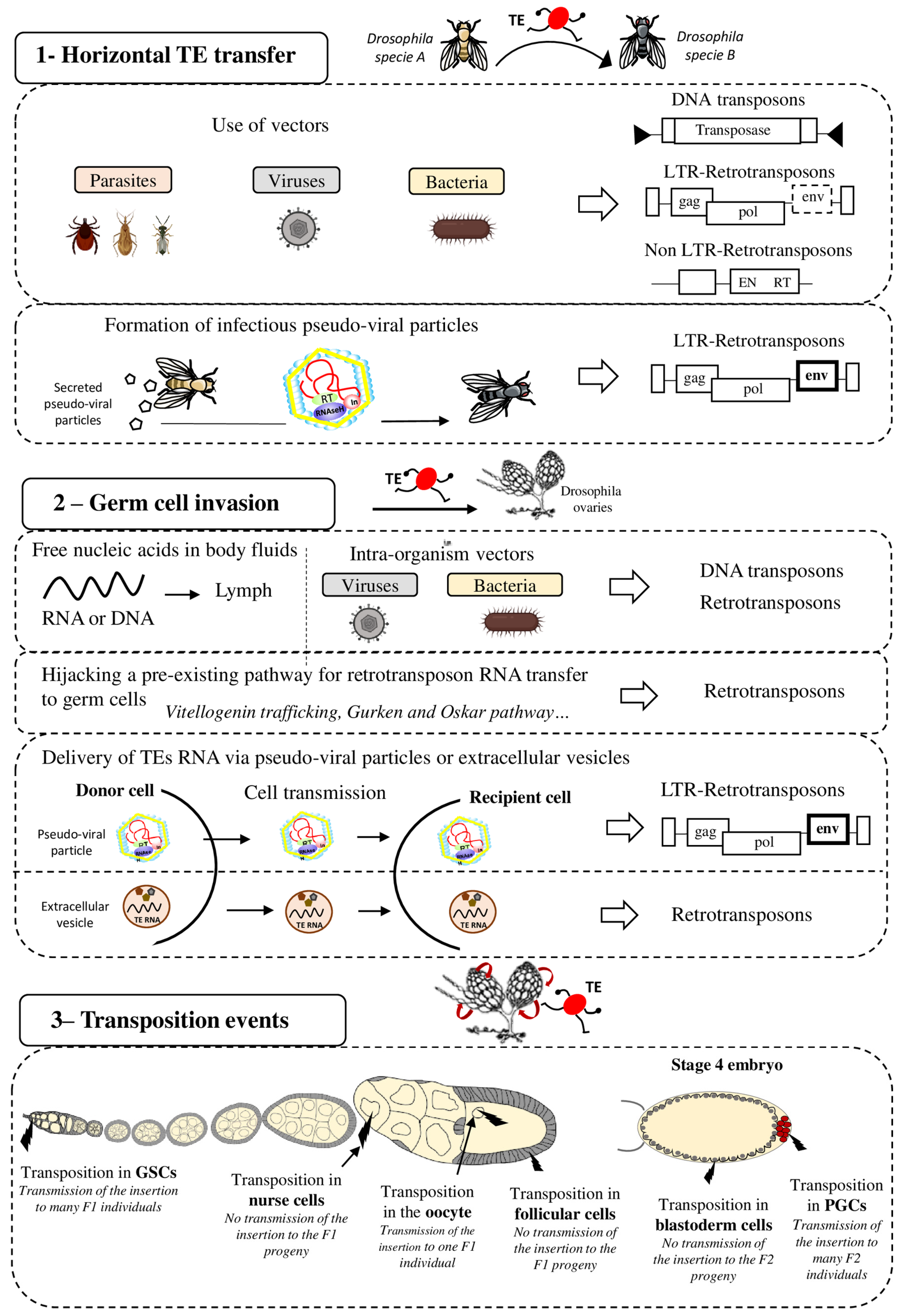
Figure 1. Strategies for effective spreading of transposable elements in the Drosophila genome. Transposable elements (TE), DNA transposons and LTR and non-LTR retrotransposons, can colonize “naive” genomes through horizontal transfer (panel 1). This may occur via vectors (e.g., parasites, viruses, and bacteria) that transfer genetic content from one organism to another. It has also been proposed that LTR retrotransposons (ERV) can form pseudo-viral particles with infectious properties. Then, invading TEs need to reach the host’s germline (panel 2) for vertical transmission to the progeny. To this end, TE sequences (excised DNA or RNA for retrotransposons) might circulate in the blood or be transported to germ cells by intra-organism vectors (viruses, bacteria). Retrotransposons can hijack pre-existing cell pathways, such as the vitellogenin trafficking (ZAM), Gurken (I-element) or Oskar (TAHRE) pathways, to transfer their RNA to germ cells. LTR-retrotransposons (ERV), through the formation of infectious pseudo-viral particles, might also be transferred between cells and/or circulate through extracellular fluids to reach the host’s germline. Extracellular vesicles have also been proposed to be efficient vectors for transferring TEs between host cells The last step for efficient invasion requires transposition in germ cells (panel 3) that allows or not (in function of the cell type) the TE transmission and propagation in the specie. GSC = Germinal Stem Cells, PGC = Primordial Germ Cells.
2.2. Spread within an Organism and Vertical Transfer to Descendants
2.2.1. Germ Cell Invasion: A Life or Death Issue
After invasion of an organism, TEs must then reach the host’s germ cells, the only cells whose genetic material will be transmitted to the offspring (vertical transmission) (Figure 1 panel 2). Indeed, if transposition occurs only in somatic cells, the horizontally transferred TEs will die with the host and will never spread within that species. Therefore, successful HTT requires the TE integration into the germ cell genome. Although germline and soma are well distinct cell types in animals, HTT is not rare in complex multicellular eukaryotes, showing that genetic material is transferred from soma to germline. It is still unknown when and in which cells during germ cell development TEs transpose and integrate the germline genome after HTT. Indeed, TE sequences must circulate in the body to reach the germline. They may be transported by their own pseudo-viral particles or by vectors, such as Wolbachia bacteria, or even in the form of free RNA or DNA [25]. DNA and RNA molecules can circulate in the extracellular body fluids, such as blood, plasma, lymph, saliva, or milk [26]. It has also been proposed that extracellular vesicles (EVs) could be efficient vectors for transferring TEs between host cells [27]. EVs, such as exosomes or microvesicles, are cell-derived vesicles that deliver biological molecules between different cells and cell types in organisms [28]. LINE1 retrotransposon RNAs have been detected in EVs isolated from cells expressing LINE1 active elements, and these EVs can then deliver LINE1 RNA to recipient cells. Thus, EVs could deliver retrotransposon RNA to neighboring and distant cells, potentially permitting germ cell invasion.
Once the TE reaches the germ cells, transposition into the germ cell genome is the next critical step to ensure its inheritance. It is important to note that gonads are made of different cell types and that transposition can occur at different stages of gametogenesis and in the different cell types (Figure 1 panel 3). The consequences of transposition depend on the type of gonad cells in which transposition occurs. For instance, in Drosophila embryos, Primordial Germ Cells (PGCs) give rise to all Germinal Stem Cells (GSCs) present in adult gonads. In adults, GSCs divide asymmetrically to produce one daughter GSC and one cystoblast. The cystoblast begins to differentiate and undergoes four rounds of mitotic divisions to form a cyst of 16 germinal cells. Most of them become nurse cells and only one will differentiate into the future oocyte. The oocyte is the only germ cell that will progress through meiosis and will be fertilized. Nurse cells do not transfer their genetic content to the progeny. Consequently, transposition in nurse cells is not expected to be beneficial to the TE. However, these cells show a high level of polyploidy and produce huge RNA quantities. For TEs, nurse cell invasion could be an intermediate and suitable strategy to reach the oocyte, especially if they transpose through an RNA intermediate, as retrotransposons do. Once inserted in the DNA of nurse cells, TEs might be expressed and produce a huge quantity of RNA that will then be transmitted to the oocyte when nurse cells dump their content into the oocyte. In agreement, the I-element (a Drosophila retrotransposon) is expressed only in nurse cells and then I-element RNA transits to the oocyte to integrate its genome [29]. The RNAs of other TEs (HMS-Beagle, 3S18, Blood, Max, TAHRE, Burdock, and HeT-A) also can target the oocyte [29][30][31]. TEs have developed a variety of strategies to optimize their transfer to specific target cells, such as the oocyte, particularly by hijacking pre-existing pathways in the host organism. For instance, I-elements target the oocyte nucleus by exploiting the host transport machinery of gurken mRNA [32]. I-element transcripts contain a small loop of secondary structure that resembles a structure present in gurken mRNA. This loop represents a consensus signal for targeting RNAs to the oocyte nucleus by dynein-mediated transport. Similarly, TAHRE transcripts migrate to the oocyte germ plasm by mimicking oskar RNAs and engaging the Staufen-dependent active transport machinery [30]. Once in the oocyte, TE RNA can be reverse transcribed before integration in the oocyte DNA. However, the highly condensed oocyte genome makes transposition events difficult and unlikely in this cell type. Yet, RNAs present in the ooplasm are transmitted to the embryo. Thus, transposition can also occur in the embryo, particularly in its PGCs. TE integration in the embryo PGCs is particularly advantageous because the new insertions will be inherited by the next generation through the myriad of gametes produced. For example, P-elements can transpose to PGCs in embryos and in GSCs in adult ovaries, and this could explain their rapid propagation in D. melanogaster populations [33].
2.2.2. Last Step for an Efficient Invasion: Transposition and Fixation in Target Cells
Transposition in the germ cell genome is crucial for TE propagation in a population because it allows the vertical transmission of new insertions. However, the transposition rate is very low. For example, 10−4 transposition events per TE copy per generation occur in Drosophila natural and laboratory populations [34][35][36]. Although the transposition rate can be higher in conditions of environmental or genomic stress, this is not sufficient to explain how genome invasion is quickly observed after HTT. According to the model proposed by Le Rouzic and Capy, following invasion by HT, an initial transposition burst occurs [37] that leads to TE accumulation in the genome before the induction of an adaptive response by the host to control transposition. Unfortunately, it is almost impossible to observe transposition bursts in real time because they can be very fast. However, the rapid invasion of P-elements in natural D. melanogaster populations following HTT strongly suggests that the rate of P-element transposition must have been very high at one point. Interestingly, Kofler and al. have demonstrated that D. simulans populations that are at their starting point of P-element invasion have a high P-element transposition rate during the first time of invasion [38]. A similar observation has been made analyzing the transposition dynamics of the mariner DNA transposon after its introduction into D. melanogaster populations containing no active mariner [39]. The high transposition rate of the introduced mariner element leads to the invasion of the population and the colonization of the genome. Finally, by artificially introducing TEs in naive species to mimic HTT, it has been demonstrated that the introduction of DNA transposons (e.g., Tc1, hAT, and PiggyBag) in the genome of species that belong to different kingdoms or domains of life leads to high transposition rates that depend on the TE class and expression pattern [40]. It is important to note that the capacity to transpose may vary among TEs. As an example, it has been proposed that the great success of DNA transposons to transfer horizontally compared to retrotransposons could be explained by their “blurry promoters” [41]. Actually, DNA transposon expression shows very low dependence on host factors; these TEs are more broadly expressed in diverse organisms allowing them to transpose in a large panel of hosts. Another explanation could be that DNA intermediate molecules of DNA transposons are more stable than RNA intermediate molecules used for retrotransposon transposition.
At the end, TEs that can invade genomes are certainly those that can implement an efficient invasion strategy and transposition mechanisms. Moreover, insertions that are neutral or that increase the host fitness have higher chances to be fixed in a population [42]. However, a quantitative population genetics model showed that a TE may persist as long as its deleterious effect on the host is lower than the advantage of transposition explaining that even TEs with negative fitness effects may spread in populations [43][44].
This led to the conclusion that selective pressure is exerted on TEs during the first steps of invasion, depending on their burst capacity and the effect of TE insertions on population fitness. However, there is a “common advantage” for both host and TEs in limiting massive transposition in the whole organism. In fact, transposition in somatic cells can be deleterious by creating detrimental mutations that lead to the host death and concomitantly to the TE disappearance. From this point of view, TEs resemble viruses: they must multiply and spread, but the host also must survive. It has been hypothesized that several TEs, such as the P-element and the I-element, have developed the ability to be expressed only in germ cells [45][46][47]. This avoids the deleterious effects of mutations in somatic cells and ensures the transmission of new TE insertions to the progeny. However, transposition in germ cells could also have dangerous outcomes because these mutations are inheritable. Transposition in PGCs (the precursors of all germ cells) is certainly very efficient for TE invasion, but it is highly risky. Indeed, germline transpositions may induce infertility and also deleterious inheritable pathology-causing mutations that potentially endanger the species’ survival. One of the best described deleterious effects of TE transposition in the germline concerns the massive mobilization of TEs that might have contributed to the extinction of Wrangel island mammoths. This small population of mammoths accumulated a large number of detrimental mutations, including deletions and point mutations, and also many TE sequences. This suggests high TE activity in the mammoth germline that led to a very high number of heritable mutations. This high transposition rate in the germline may have contributed, with other factors, to the extinction of this small endangered population [48].
Therefore, TE transposition must be controlled in all cell types to limit its negative effects on the host and its progeny. On the other hand, a too strict control can cause the TE loss from the host genome and deprive the organism of an important source of genetic diversity. For example, Spermophilus tridecemlineatus is a rodent in which transposon activity has declined over at least 4 million years. Its genome does not harbor any recent TE, Long Interspersed Nucleotide Element (LINE), SINE, retrotransposon with long terminal repeats (LTR), or DNA transposon activity. Moreover, no functional TE copy is found in the genome of this species because all harbor a huge number of mutations. This is explained by the strong TE silencing, leading to complete inhibition of TE mobilization [49]. To be conserved in a genome, a minimum of transposition is required. Interestingly, in several eukaryotes, temporary relaxation of the TE silencing machinery has been observed in the germline and its associated cells. For instance, during Drosophila early oogenesis, there is a short spatiotemporal window when the piRNA pathway seems to be less efficient and at least some TEs might escape the host control. It has been proposed that this window, termed the ‘Piwiless pocket’, allows the insertion of new TEs in the developing germline genome [50][51][52]. In mammals, transient TE relaxation during germ cell development has been observed mainly during epigenetic reprogramming periods [5][53]. Specifically, during the first wave of global reprograming that occurs following fertilization, 10% of the transcriptome in 2-cell stage mouse embryos is made of specific TE transcripts, including transcripts from the MuERV retroelement [54][55][56]. The second reprogramming wave occurs in PGCs of the developing mouse embryo. Although no general transcriptional burst has been observed for TEs at this step, some specific TE transcripts (i.e., LINE1 transcripts) are overrepresented [53], reviewed in [57]. The presence of such spatiotemporal windows during germline development in which TE control is weaker could help to explain the very successful genome invasion by TEs. Once settled in the germline genome, new TE insertions are then vertically transmitted, like any other DNA sequence.
2.3. Retrotransposons: A Formidable Capacity of Propagation
2.3.1. Retrotransposons Can Do Intercellular Transposition
Retrotransposons have evolved in a variety of organisms, from protozoa to humans, and display outstanding capacities of rapid invasion and propagation. There are two types of retrotransposons: with and without LTR. Non-LTR retrotransposons lack LTR and have generally two open reading frames of which one encodes a reverse transcriptase and an endonuclease. SINEs do not encode a functional reverse transcriptase and are non-autonomous elements because their transposition relies on enzymes encoded by other non-LTR retrotransposons: the LINEs. Thus, SINEs cannot colonize a naive genome after HT if the genome does not have a corresponding element for trans-complementation.
The other retrotransposon group: LTR retrotransposons. LTR retrotransposons resemble retroviral proviruses. Indeed, they have LTRs at each extremity and open reading frames equivalent to the gag and pol genes. Gag encodes a structural protein involved in the formation of virus-like particles. Pol encodes proteins that are necessary for transposition mechanisms: an integrase, a RNase H, a protease, and a reverse transcriptase. Some LTR retrotransposons also harbor the envelope gene (env) that encodes a viral surface glycoprotein, and they are called endogenous retroviruses (ERV). ERVs are assumed to be derived from past retroviral infections that have been integrated as permanent residents in host genomes. Like retroviruses, most ERVs can form virus-like particles (VLP). The Env protein interacts with target host cell receptors and allows the fusion of the VLP with the target cell membrane and ERV propagation between cells. ERVs make up approximatively 10% of the mouse, rat, and human genomes and they have been extensively studied in Drosophila [8]. In the Drosophila genome, many ERVs copies are present, such as the very diverse Gypsy-like elements including ZAM for instance. These TEs can form VLPs and infect neighboring cells [58][59].
2.3.2. Horizontal Transfer of Retrotransposons: Do They Really Need Vectors?
It has been hypothesized that VLPs produced by ERVs can propagate between organisms, like retrovirus particles, without vectors (Figure 1 panel 1). In this case, these particles could be infectious. VLPs of the Gyspy ERV have been found as extracellular particles in the medium in which D. melanogaster follicular cells were cultured. This means that Gypsy VLPs are secreted by cells [60]. Moreover, these VLPs can infect cultured cells belonging to another Drosophila species: Drosophila hydei. Interestingly, a recent study showed that Gypsy also transits between cells that are not in contact [61]. Furthermore, experiments in which flies were grown on medium containing crushed pupae that produced Gypsy VLPs suggested a possible HTT via food: these flies became infected by Gypsy [62]. Once transmitted to a new individual by HTT, retrotransposons could use their retroviral properties to propagate between cells and through body fluids to reach the germline, ensuring their spread in that species.
2.3.3. Drosophila Germ Cell Invasion by Retrotransposons
Most TEs can insert in the germline by being active directly in these cells. On the other hand, ERVs do not seem to be expressed in germ cells. In Drosophila ovaries, when the pathway regulating TEs is abolished in all cell types, ERVs are only expressed in a patch of somatic cells that are called follicular cells and that surround germ cells [63][64][65][66][67][68]. Indeed, as described for gene transcription, TE transcription requires transcription factors that are present in specific cell subsets [69][70]. For instance, ZAM retrotransposon expression requires the presence of Pointed 2, a transcription factor only expressed in a patch of follicular cells [70]. This means that ERV RNAs are not produced directly in germ cells and that transposition into the germ cell genome requires ERV transmission from somatic cells. VLPs formed in the producing somatic cells could infect germ cells via the Env transmembrane protein, but other routes could also be used (Figure 1 panel 2). For instance, the 412 element can infect germ cells, although it does not encode Env [71]. This retrotransposon might use the Env protein encoded by another ERV for germ cell infection. Moreover, the ZAM ERV encodes an Env protein, but it reaches germ cells by usurping the endosome/exosome pathway in Drosophila ovaries for VLP transfer to the oocyte [59]. This route is normally employed for vitellogenin release and uptake by germ cells. It is not known whether ZAM also uses its Env protein to transit to germ cells. The detection of many new ZAM insertions in the progeny of flies in which ZAM is derepressed in somatic follicular cells indicates that after VLP transfer, ZAM can insert into the germ cells genome [72][73]. Therefore, this mechanism of propagation, using transfer from somatic to germ cells, is an efficient way to spread in a population. This suggests that ERVs expressed in somatic cells close to germ cells might create particles that infect the germline or might use a more passive mechanism to reach germ cells for insertion in the genome and vertical transmission.
It is clear that even if most ERVs are not expressed in the germline, they have many strategies for efficient propagation. Thanks to their capacity of intercellular and potentially inter-organism transfers, ERVs seem particularly well suited to efficiently propagate in an organism, to its descendants, and also to other species. This could explain why retrotransposons occupy such an important place in eukaryote genomes [8].
This entry is adapted from the peer-reviewed paper 10.3390/biology11050710
References
- McClintock, B. Induction of Instability at Selected Loci in Maize. Genetics 1953, 38, 579–599.
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982.
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103.
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science 2007, 315, 1587–1590.
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdžić, M.; Becker, J.D.; Feijó, J.A.; Martienssen, R.A. Epigenetic Reprogramming and Small RNA Silencing of Transposable Elements in Pollen. Cell 2009, 136, 461–472.
- Wolf, D.; Goff, S.P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 2009, 458, 1201–1204.
- Wolf, G.; Yang, P.; Füchtbauer, A.C.; Füchtbauer, E.-M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015, 29, 538–554.
- Huang, C.R.L.; Burns, K.H.; Boeke, J.D. Active Transposition in Genomes. Annu. Rev. Genet. 2012, 46, 651–675.
- Zeh, D.W.; Zeh, J.A.; Ishida, Y. Transposable elements and an epigenetic basis for punctuated equilibria. BioEssays 2009, 31, 715–726.
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703.
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368.
- Oliver, K.R.; Greene, W.K. Transposable elements: Powerful facilitators of evolution. BioEssays 2009, 31, 703–714.
- Daniels, S.B.; Peterson, K.R.; Strausbaugh, L.D.; Kidwell, M.G.; Chovnik, A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 1990, 124, 339–355.
- Anxolabéhère, D.; Nouaud, D.; Périquet, G.; Tchen, P. P-element distribution in Eurasian populations of Drosophila melanogaster: A genetic and molecular analysis. Proc. Natl. Acad. Sci. USA 1985, 82, 5418–5422.
- Anxolabéhère, D.; Kidwell, M.G.; Periquet, G. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 1988, 5, 252–269.
- Ghanim, G.E.; Rio, D.C.; Teixeira, F.K. Mechanism and regulation of P element transposition. Open Biol. 2020, 10, 200244.
- Peccoud, J.; Loiseau, V.; Cordaux, R.; Gilbert, C. Massive horizontal transfer of transposable elements in insects. Proc. Natl. Acad. Sci. USA 2017, 114, 4721–4726.
- Houck, M.A.; Clark, J.B.; Peterson, K.R.; Kidwell, M.G. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science 1991, 253, 1125–1128.
- Loreto, E.L.S.; Carareto, C.M.A.; Capy, P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity 2008, 100, 545–554.
- Gilbert, C.; Peccoud, J.; Chateigner, A.; Moumen, B.; Cordaux, R.; Herniou, E.A. Continuous Influx of Genetic Material from Host to Virus Populations. PLoS Genet. 2016, 12, e1005838.
- Piskurek, O.; Okada, N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 12046–12051.
- Dunning Hotopp, J.C.; Clark, M.E.; Oliveira, D.C.S.G.; Foster, J.M.; Fischer, P.; Muñoz Torres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.; et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756.
- Raychoudhury, R.; Baldo, L.; Oliveira, D.C.S.G.; Werren, J.H. Modes of acquisition of Wolbachia: Horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evol. Int. J. Org. Evol. 2009, 63, 165–183.
- Palazzo, A.; Escuder, E.; D’Addabbo, P.; Lovero, D.; Marsano, R.M. A genomic survey of Tc1-mariner transposons in nematodes suggests extensive horizontal transposon transfer events. Mol. Phylogenet. Evol. 2021, 158, 107090.
- White, P.M.; Pietri, J.E.; Debec, A.; Russell, S.; Patel, B.; Sullivan, W. Mechanisms of Horizontal Cell-to-Cell Tranfer of Wolbachia spp. in Drosophila melanogaster. Appl. Environ. Microbiol. 2017, 83, e03425-16.
- Stroun, M.; Lyautey, J.; Lederrey, C.; Mulcahy, H.E.; Anker, P. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: Evidence for a preferential release from viable cells? Ann. N. Y. Acad. Sci. 2001, 945, 258–264.
- Kawamura, Y.; Sanchez Calle, A.; Yamamoto, Y.; Sato, T.A.; Ochiya, T. Extracellular vesicles mediate the horizontal transfer of an active LINE-1 retrotransposon. J. Extracell. Vesicles 2019, 8, 1643214.
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232.
- Wang, L.; Dou, K.; Moon, S.; Tan, F.J.; Zhang, Z.Z. Hijacking Oogenesis Enables Massive Propagation of LINE and Retroviral Transposons. Cell 2018, 174, 1082–1094.e12.
- Tiwari, B.; Kurtz, P.; Jones, A.E.; Wylie, A.; Amatruda, J.F.; Boggupalli, D.P.; Gonsalvez, G.B.; Abrams, J.M. Retrotransposons Mimic Germ Plasm Determinants to Promote Transgenerational Inheritance. Curr. Biol. 2017, 27, 3010–3016.e3.
- Shpiz, S.; Kwon, D.; Uneva, A.; Kim, M.; Klenov, M.; Rozovsky, Y.; Georgiev, P.; Savitsky, M.; Kalmykova, A. Characterization of Drosophila telomeric retroelement TAHRE: Transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 2007, 24, 2535–2545.
- Van De Bor, V.; Hartswood, E.; Jones, C.; Finnegan, D.; Davis, I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev. Cell 2005, 9, 51–62.
- Moon, S.; Cassani, M.; Lin, Y.A.; Wang, L.; Dou, K.; Zhang, Z.Z. A Robust Transposon-Endogenizing Response from Germline Stem Cells. Dev. Cell 2018, 47, 660–671.e3.
- Suh, D.S.; Choi, E.H.; Yamazaki, T.; Harada, K. Studies on the transposition rates of mobile genetic elements in a natural population of Drosophila melanogaster. Mol. Biol. Evol. 1995, 12, 748–758.
- Nuzhdin, S.V.; Mackay, T.F. The genomic rate of transposable element movement in Drosophila melanogaster. Mol. Biol. Evol. 1995, 12, 180–181.
- Maside, X.; Assimacopoulos, S.; Charlesworth, B. Rates of movement of transposable elements on the second chromosome of Drosophila melanogaster. Genet. Res. 2000, 75, 275–284.
- Le Rouzic, A.; Capy, P. The first steps of transposable elements invasion: Parasitic strategy vs. genetic drift. Genetics 2005, 169, 1033–1043.
- Kofler, R.; Senti, K.A.; Nolte, V.; Tobler, R.; Schlötterer, C. Molecular dissection of a natural transposable element invasion. Genome Res. 2018, 28, 824–835.
- Robillard, É.; Le Rouzic, A.; Zhang, Z.; Capy, P.; Hua-Van, A. Experimental evolution reveals hyperparasitic interactions among transposable elements. Proc. Natl. Acad. Sci. USA 2016, 113, 14763–14768.
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 2010, 25, 537–546.
- Zhang, H.H.; Peccoud, J.; Xu, M.R.X.; Zhang, X.G.; Gilbert, C. Horizontal transfer and evolution of transposable elements in vertebrates. Nat. Commun. 2020, 11, 1362.
- Capy, P. Taming, Domestication and Exaptation: Trajectories of Transposable Elements in Genomes. Cells 2021, 10, 3590.
- Hickey, D.A. Selfish DNA: A sexually transmitted nuclear parasite. Genetics 1982, 101, 519–531.
- Kofler, R. Dynamics of transposable element invasions with piRNA clusters. Mol. Biol. Evol. 2019, 36, 1457–1472.
- Laski, F.A.; Rio, D.C.; Rubin, G.M. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 1986, 44, 7–19.
- Chaboissier, M.C.; Busseau, I.; Prosser, J.; Finnegan, D.J.; Bucheton, A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990, 9, 3557–3563.
- del Carmen Seleme, M.; Disson, O.; Robin, S.; Brun, C.; Teninges, D.; Bucheton, A. In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic Acids Res. 2005, 33, 776–785.
- Rogers, R.L.; Slatkin, M. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 2017, 13, e1006601.
- Platt, R.N.; Ray, D.A. A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene 2012, 500, 47–53.
- Dufourt, J.; Vaury, C. During a short window of Drosophila oogenesis, piRNA biogenesis may be boosted and mobilization of transposable elements allowed. Front. Genet. 2014, 5, 8–11.
- Théron, E.; Maupetit-Mehouas, S.; Pouchin, P.; Baudet, L.; Brasset, E.; Vaury, C. The interplay between the argonaute proteins piwi and aub within drosophila germarium is critical for oogenesis, piRNA biogenesis and TE silencing. Nucleic Acids Res. 2018, 46, 10052–10065.
- Dufourt, J.; Dennis, C.; Boivin, A.; Gueguen, N.; Théron, E.; Goriaux, C.; Pouchin, P.; Ronsseray, S.; Brasset, E.; Vaury, C. Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis. Nucleic Acids Res. 2014, 42, 2512–2524.
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Mol. Cell 2012, 48, 849–862.
- Evsikov, A.V.; De Vries, W.N.; Peaston, A.E.; Radford, E.E.; Fancher, K.S.; Chen, F.H.; Blake, J.A.; Bult, C.J.; Latham, K.E.; Solter, D.; et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004, 105, 240–250.
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013, 20, 332–338.
- Kigami, D.; Minami, N.; Takayama, H.; Imai, H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 2003, 68, 651–654.
- Maupetit-Mehouas, S.; Vaury, C. Transposon Reactivation in the Germline May Be Useful for Both Transposons and Their Host Genomes. Cells 2020, 9, 1172.
- Chalvet, F.; Teysset, L.; Terzian, C.; Prud’homme, N.; Santamaria, P.; Bucheton, A.; Pélisson, A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999, 18, 2659–2669.
- Brasset, E.; Taddei, A.R.; Arnaud, F.; Faye, B.; Fausto, A.M.; Mazzini, M.; Giorgi, F.; Vaury, C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 2006, 3, 25.
- Syomin, B.V.; Fedorova, L.I.; Surkov, S.A.; Ilyin, Y.V. The endogenous Drosophila melanogaster retrovirus gypsy can propagate in Drosophila hydei cells. Mol. Gen. Genet. 2001, 264, 588–594.
- Keegan, R.M.; Talbot, L.R.; Chang, Y.H.; Metzger, M.J.; Dubnau, J. Intercellular viral spread and intracellular transposition of Drosophila gypsy. PLoS Genet. 2021, 17, e1009535.
- Kim, A.; Terzian, C.; Santamaria, P.; Pélisson, A.; Prud’homme, N.; Bucheton, A. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1994, 91, 1285–1289.
- Leblanc, P.; Desset, S.; Giorgi, F.; Taddei, A.R.; Fausto, A.M.; Mazzini, M.; Dastugue, B.; Vaury, C. Life Cycle of an Endogenous Retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 2002, 74, 10658–10669.
- Tcheressiz, S.; Calco, V.; Arnaud, F.; Arthaud, L.; Dastugue, B.; Vaury, C. Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol. Genet. Genomics 2002, 267, 133–141.
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 2009, 137, 522–535.
- Olivieri, D.; Sykora, M.M.; Sachidanandam, R.; Mechtler, K.; Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010, 29, 3301–3317.
- Pélisson, A.; Song, S.U.; Prud’homme, N.; Smith, P.A.; Bucheton, A.; Corces, V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994, 13, 4401–4411.
- Sokolova, O.A.; Mikhaleva, E.A.; Kharitonov, S.L.; Abramov, Y.A.; Gvozdev, V.A.; Klenov, M.S. Special vulnerability of somatic niche cells to transposable element activation in Drosophila larval ovaries. Sci. Rep. 2020, 10, 1076.
- Cavarec, L.; Jensen, S.; Casella, J.F.; Cristescu, S.A.; Heidmann, T. Molecular cloning and characterization of a transcription factor for the copia retrotransposon with homology to the BTB-containing lola neurogenic factor. Mol. Cell. Biol. 1997, 17, 482–494.
- Meignin, C.; Dastugue, B.; Vaury, C. Intercellular communication between germ line and somatic line is utilized to control the transcription of ZAM, an endogenous retrovirus from Drosophila melanogaster. Nucleic Acids Res. 2004, 32, 3799–3806.
- Barckmann, B.; El-Barouk, M.; Pélisson, A.; Mugat, B.; Li, B.; Franckhauser, C.; Fiston Lavier, A.-S.; Mirouze, M.; Fablet, M.; Chambeyron, S. The somatic piRNA pathway controls germline transposition over generations. Nucleic Acids Res. 2018, 46, 9524–9536.
- Duc, C.; Yoth, M.; Jensen, S.; Mouniée, N.; Bergman, C.M.; Vaury, C.; Brasset, E. Trapping a somatic endogenous retrovirus into a germline piRNA cluster immunizes the germline against further invasion. Genome Biol. 2019, 20, 127.
- Desset, S.; Conte, C.; Dimitri, P.; Calco, V.; Dastugue, B.; Vaury, C. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 1999, 16, 54–66.
This entry is offline, you can click here to edit this entry!
