Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Polymers including guar gum and related compounds, cellulose-based gels, and acrylamide-based polymers (PAM and HPAM) are used in the petroleum industry mainly for increasing viscosity and reducing friction in the reservoirs during water flooding and hydraulic fracturing.
- polymer
- biodegradation
- enzyme biotechnology
1. Guar Gum
Guar gum is typically extracted from the seed of the guar plant, Cyamopsis tetragonolobus, and it is widely used in many applications, particularly in the food industry [1]. Guar and its derivatives are also commonly used in hydraulic fracturing processes, although it has been reported that the polymer is sensitive to prolonged heat and as such does not keep its rheological properties [2]. Guar is a polysaccharide consisting of mannose units, bound together by β-1,4 glyosidic bonds and randomly attached galactose molecules linked to the mannose backbone by α-1,6 linkages [3]. The ratio of mannose compared to galactose units is usually between 1.8:1 and 2:1, and depends on the provenance of the polymer [1]. The rheological properties of guar gum depend on the length of the backbone and the mannose:galactose ratio [4]. The biodegradation of guar gum is catalyzed by hydrolases that attack the β-1,4 and α-1,6 linkages, by β-1,4-mannanase, β-mannosidase and α-1,6-galactosidase, resulting in its degradation into simple monosaccharides and disaccharides [5]. The β-1,4-mannanase cleaves the mannosidic bonds, having a direct effect on the viscosity of a guar solution as β-mannosidase act on the ends of the polymer chain and hydrolyzes the terminal glycoside group of guar gum. The α-galactosidase is responsible for the removal of the galactosidase units [4].
Guar-linkage specific enzymes (GLS) have been investigated for their use as enzyme breakers in hydraulic fracturing processes at different pH ranges, temperatures, and salinities. For example, GLS enzymes isolated from the fungus Aspergillus niger were shown to effective break (degrade) guar at temperatures between 15 to 60 °C, and at pH values of 3 to 11 [6]. Thermostable α-1,6-galactosidase and β-1,4-mannanase enzymes were isolated from the hyperthermophile Thermotoga neapolitana, and were shown to be active at temperatures of up to 100 °C [5]. Similarly, Thermotoga maritima was reported for its expression of a thermostable α-galactosidase enzyme at 85 °C [7]. Another thermophilic bacterium isolated from hot springs, Rhodothermus marimus, showed galactomannan degradation activity by expressing a mannanase having an optimal activity at 85 °C and a pH of 5.4 [8]. A lower temperature (50 °C) mannanase isolated from Enterobacter sp. has also been reported to be an effective enzyme breaker for guar gum at a pH range of 3.0 to 8.0, and at high salt concentrations (up to 4 M NaCl) [9]. Fridjonsson et al. [10] also reported an α-galactosidase from the genus Thermus brockianus ITI360 cloned into E. coli that was active at an optimal pH of 5.5 to 6.5 and optimal temperature of 93 °C. More recently, a protein-engineered galacto-mannanase was identified that was able to degrade the polymer up to 120 °C, which is the highest recorded temperature at which enzymes were active to degrade guar [11]. Similarly, the enzymes α-amylase and β-glucanase that are able to degrade xanthan gum and starch-based polymers, have also been found to be effective at temperatures up to 90 °C [12]. The long-term effectiveness of GLS enzymes to degrade guar-based filter cakes has been tested in oilfields relative to persulfate oxidizers, summarized by Brannon et al. [13].
2. Cellulose-Based Polymers
Due to the fluctuation in price and occasional supply shortage of guar gum, other gelling agents are now often added to hydraulic fracturing solutions, including cellulose-based polymers such CMC and carboxymethylhydroxyethylcellulose (CMHEC) [14]. Azizov et al. [ 57] showed that the use of CMC in fracturing fluid systems can result in similar production performance and lower cost relative to guar gum. With increased industry interest in using polymers such as CMC in hydraulic fracturing operations, understanding the biodegradation of cellulose-based polymers is important for developing new enzyme breakers against these types of filter cakes.
Cellulose is a polymer that consists of repeating units of glucose linked by β-1,4 bonds [15] and CMC consists of a cellulose molecule with random carboxymethyl groups replacing hydroxyl groups within the molecule [16]. The chemical reaction for the replacement of the hydroxyl groups involves an alkali-catalyzed reaction with chloroacetic acid. The carboxymethyl groups render CMC soluble and chemically reactive [17]. The biodegradation of CMC occurs mostly via a cellulose-enzyme complex, known as a cellulosome, that includes: exo-β-1,4-glucanases (1), endo-β-1,4-glucanases (2) and β-1,4-glucosidases (3) [18]. Cellulosomes are complexes of cellulases bound to different scaffolding proteins such as carbohydrate-binding modules, docking modules, cohesion modules, and surface layer homology modules [19]. Exoglucanases attack the end of the CMC molecule, resulting in glucose or cellobiose formation, endoglucanases break down internal glucosidic bonds, and glucosidases catalyze the hydrolysis of cellobiose, forming glucose [18][19]. Although many anaerobic bacteria express these multi-protein complexes, some anaerobes also hydrolyze cellulose or related molecules by expressing a single enzyme [20]. As the action of endoglucanases lowers the molecular weight of CMC, thus decreasing its viscosity, these enzymes are the ideal candidates for degrading the CMC-based filter cakes in oil reservoirs.
The production of endoglucanases from both fungi [21] and bacterial species [22] have found widespread application in the food and agricultural industries. In contrast, comparatively few studies have examined the development of endoglucanases for applications in the petroleum energy industry. Therefore, there is a limited understanding of the activity of endoglucanases under the environmental conditions that characterize subsurface petroliferous reservoirs, such as low redox conditions, and high salinities, temperatures, and pressures. While enzyme breakers have been developed to degrade cellulose-based polymers at temperatures between 15 to 60 °C and pH between 1 to 8 [14][23], most studies on the biodegradation of CMC filter cakes have been conducted with purified enzymes. However, their properties and identities are usually kept confidential (e.g., reported in patents) [24][25][26], thus limiting the progress in CMC enzyme breaker technology development. Trabelsi et al. [27] observed a viscosity decrease in guar and CMC when two different enzymes were tested as breakers at low pH (4.75), and at a relatively high temperature (49 °C). However, the protein sequences of the enzymes, nor their microbial origins, were reported [27]. Recently, CMC-degrading enzymes were retrieved from a thermophilic (50 °C), methanogenic enrichment culture established from cattle manure that was supplemented with CMC as its sole carbon and energy source. Extracellular enzymes degrading CMC were able to completely hydrolyze the polymer under high temperatures (50 to 80 °C), high salinities (up to 20% (w/v) salts), and were active between pH 5 to 8 [28]. Additionally, these enzymes could reduce CMC viscosity under high pressures (up to 4000 psi). The CMC-degrading enzymes from this anaerobic culture were subsequently isolated and purified for further study and testing [29]. These latter two studies showed that CMC-degrading enzymes can potentially be used as filter cake breakers under realistic oil field conditions characterized by high salinities and temperatures, though scale up and field tests are still required.
Recent advances in proteomics have helped understanding the structure and function of cellulosome complexes which can be used for cellulose biodegradation by some anaerobic bacteria [19][20]. New techniques to isolate proteins involved in the cellulosome complex have been developed from studying the structure of the complex. Work done by Hong et al. [30] reported a new technique to isolate and purify cellulases based on the affinity of the carbohydrate-binding module to amorphous cellulose. Han et al. [31] also recently surveyed different genetic modifications such as directed evolution or chemical modifications that can be done on thermo-stable enzymes to increase their efficacy at degrading their substrates in conditions that are considered more extreme. After engineering the enzyme, its thermal stability increased such that the enzyme was active at 55 °C for 30 min and was more stable at a wider pH range (4.4 to 8.8). The interest in using CMC as an alternative to guar gum as a gelling agent in the past years [23][27] and the recent advances in proteomics and genetics offer a great opportunity to increase the research on this topic such that cellulose-based enzyme breakers can be reliably applied in oil recovery field operations.
3. PAM and HPAM
The use of non-hydrolyzed and hydrolyzed polyacrylamide (PAM and HPAM, respectively) in hydraulic fracturing fluids has increased within the last 5 years, especially in North America [32]. Therefore, it is important to understand the biodegradation of PAM and HPAM in order to develop potential enzymes that can be used as breakers to treat these types of polymer filter cakes. PAM is a high molecular weight polymer that is synthesized by polymerization of acrylamide, either as a linear chain or as a crosslinked structure [33]. Due to its high molecular weight and stable carbon backbone, PAM has been considered relatively resistant to microbial biodegradation [34][35]. It is believed that PAM and HPAM are unable to pass through microbial cell membranes, and that their carbon skeleton is difficult to access by microorganisms [36]. Nevertheless, the microbial utilization of PAM and HPAM has been reported since the late 1990s, including by microorganisms from soil, activated sludge, and oilfield production/injection waters. The amide (-NH2) groups of PAM and HPAM can be hydrolyzed and converted into ammonium (NH4+) (Figure 1A), which can then be used as a source of nitrogen for microbial growth [36][37][38]. Both aerobic and anaerobic microorganisms have been shown to utilize PAM and HPAM as nitrogen sources [39][40][41][42][43][44]. PAM or HPAM hydrolysis is catalyzed by a specific amidase enzyme, which has been repeatedly detected in microbial cultures amended with these polymers [36][41][44][45] (Figure 1A). In addition, chemical analyses have shown that during microbial utilization of PAM, its amide groups can be converted into a carboxylic acid (COOH) [39][42][44][46][47], resulting in the formation of polyacrylate. However, the utilization of the NH2 groups from PAM/HPAM (deamination) does not lead to a decrease in the viscosity or molecular weight of the polymers as the carbon-carbon backbone is not cleaved [35], and therefore amidases are not good targets for developing enzyme breakers for PAM and HPAM.
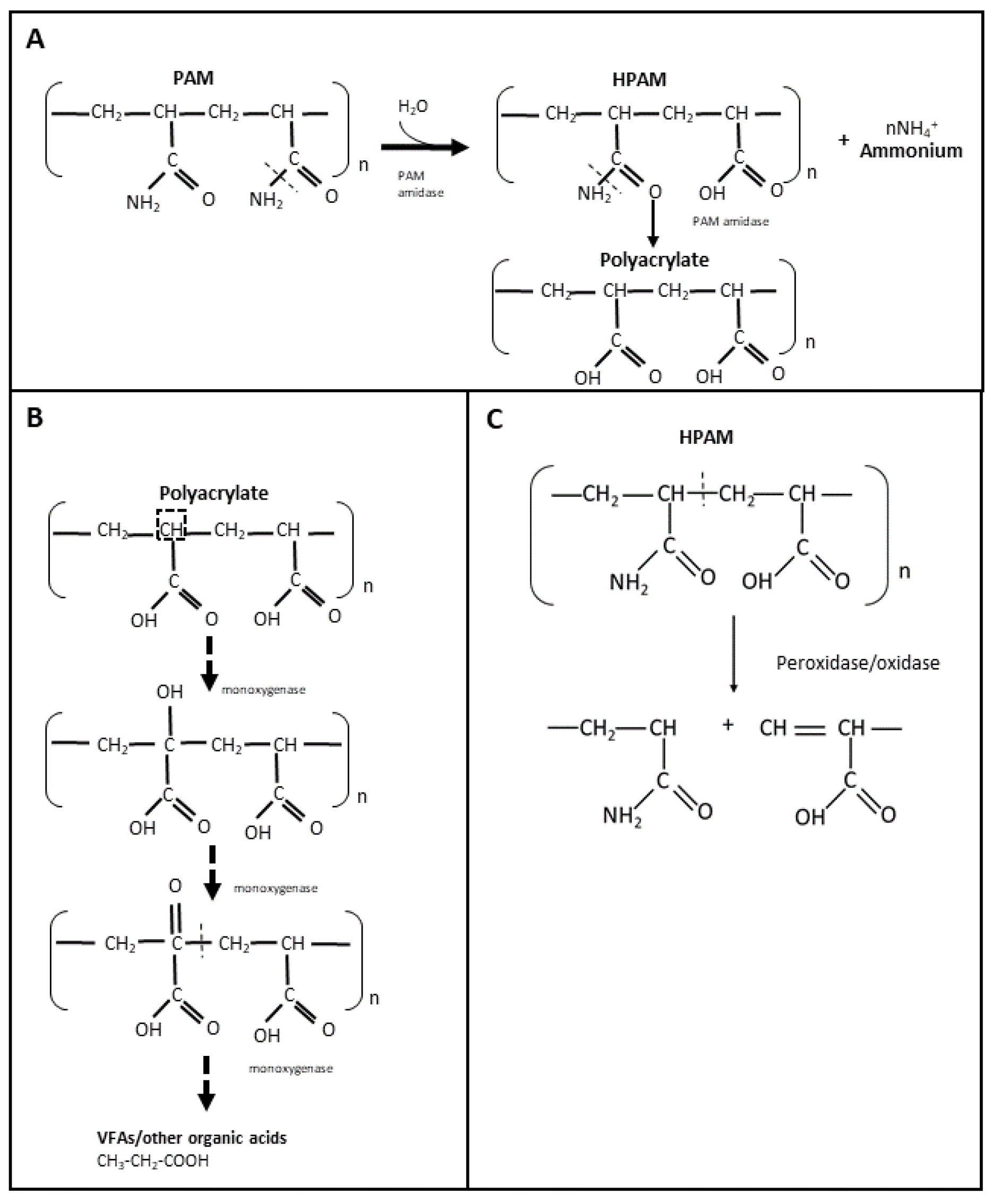
Figure 1. Microbial utilization of PAM and HPAM through hydrolyzation with amidase (A), suggested biodegradation of PAM by oxidation with monooxygenases (B), and PAM degradation mechanism by radical-forming enzymes (C).
The microbial utilization of PAM or HPAM as a carbon source is considered more challenging. Only the partial biodegradation of PAM or HPAM has been reported, although the lack of polymer-free controls in some of these studies does not unequivocally confirm if biodegradation was occurring. According to Nakamiya and Kinoshita [48], soil and activated sludge isolates degraded up to 20% of PAM after 27 h of incubation. Wen et al. [43] measured a 70% PAM removal efficiency by two Bacillus isolates after 96 h of incubation, but this degradation efficiency was assessed based on the starch-cadmium iodine assay, which measures the removal of amide groups from PAM, rather than carbon-carbon bond cleavage. Similarly, Bao et al. [39] obtained bacterial cultures from oilfield produced waters with a HPAM removal efficiency of 14%, but the cleavage of the carbon backbone in HPAM was not directly shown. More recently, microbial communities from a combined aerobic and anaerobic reactor system were shown to decrease HPAM viscosity by up to 78% [49]. In addition, the authors observed two compounds with lower molecular weight than HPAM in the aerobic system using GPC spectra [49]. Other recent studies [32][42][50][51][52] reported the presence of volatile fatty acids such as propionate, acetate and formate in microbial cultures utilizing HPAM under anaerobic conditions. Thus, it is believed these fatty acids accumulate as a result of HPAM biodegradation [32][42].
Despite the number of studies reporting PAM or HPAM biodegradation, only a few have identified specific enzymes potentially involved in the breaking of the carbon skeleton, but they should be carefully examined. Dehydrogenase and oxidases were detected in microbial systems designed for the treatment of HPAM [50][51]; however, it cannot be discerned whether these enzymes were solely produced for the purposed of HPAM biodegradation since additional carbon sources (such as glucose) were present in the systems. More recently, Song et al. [53] measured the activity of laccase (oxidase) and dehydrogenase in a combined aerobic and anaerobic bioreactor that was initially amended with glucose and urea, and then conditioned with HPAM. In this study, authors observed that laccase activity was independent of the HPAM concentration and dehydrogenase activity was indirectly proportional to the concentration of HPAM [53]. From several reports on PAM/HPAM biodegradation, it is hypothesized that the biodegradation of these polymers occurs by initial oxidation reactions that would first add a hydroxy (-OH) group into the alpha carbon of HPAM and a ketone (=O) group to allow the subsequent cleavage of the PAM/HPAM carbon skeleton, through the activity of oxygenase enzymes (e.g., monooxygenases) (Figure 1B). However, further studies are required to confirm whether these enzymes are indeed present in PAM-biodegrading cultures and if they can be used as enzyme breakers to degrade HPAM or PAM polymers in oilfield systems. A recent study added to the skepticism that HPAM or PAM polymers can be used as a carbon source [54]. Repeated transfers of microbial communities enriched from activated sludge and oilfield produced water and incubated under thermophilic conditions did not reduce the polymers’ viscosity when PAM or HPAM were provided as sole carbon sources. Instead, these polymers were shown to serve as nitrogen sources when an alternate carbon source such as glucose was provided [54].
PAM or HPAM degradation has also been reported when commercial or extracted enzymes were directly added to polymer solutions. Gupta [55] patented an enzyme breaker known as asparaginase to degrade PAM. The authors reported that this enzyme was able to deaminate the amide group of PAM and subsequently cleave the polymer, resulting in a viscosity decrease. However, the specific mechanism involved in ‘breaking’ the carbon skeleton of PAM was not shown. Other extracellular enzymes such as oxidases or peroxidases may also be effective as PAM/HPAM breakers, wherein free radicals are formed that react with the polymer carbon, leading to a cleavage in the carbon skeleton of the polymer (Figure 1C). Initially, Ramsden et al. [56] observed the degradation of a 0.5% PAM solution at 20 °C when a commercial xanthine oxidase was added in the presence of xanthine. Nakamiya et al. [57] subsequently reported the degradation of PAM when using a purified hydroquinone peroxidase enzyme isolated from Azotobacter beijerinckii HM121. In the presence of tetramethyl hydroquinone and hydrogen peroxide, this peroxidase was able to degrade PAM into polymers of smaller molecular weight within an hour of incubation at 30 °C [57]. Hydroquinone peroxidase was believed to react with hydrogen peroxide to form hydroxyl radicals which then reacted with tetramethyl hydroquinone to form a corresponding radical that attacked the carbon chain of PAM and by hydrogen abstraction broke the polymer chain [57]. Recently, Gilbert et al. [58] observed that horseradish peroxidase (HRP), in the presence of hydrogen peroxide, can also catalyze the degradation of HPAM at 37 °C by free radical formation. After 24 h, HRP decreased the viscosity and molecular weight of the HPAM solution by 81% and 67%, respectively, in the presence of 97 mM peroxide [58]. Both molecular weight and viscosity reduction were dependent on the concentration of hydrogen peroxide [58]. The results of the above studies suggest that these free radical-forming oxidases and peroxidases could potentially be used for degrading PAM or HPAM polymers in oil reservoirs, at least at mesophilic temperatures between 20 to 37 °C. However, possible interactions between the radical components formed from the potential enzymes and other chemicals present in the reservoir are yet to be investigated to confirm the effectiveness of these types of enzymes for application in oil and gas fields.
This entry is adapted from the peer-reviewed paper 10.3390/polym14091871
References
- Hasan, A.M.; Abdel-Raouf, M.E. Applications of guar gum and its derivatives in petroleum industry: A review. Egypt. J. Pet. 2018, 27, 1043–1050.
- Mudgil, D.; Barak, S.; Khatkar, B.S. Guar gum: Processing, properties and food applications-A Review. J. Food Sci. Technol. 2014, 51, 409–418.
- McCleary, B.V.; Clark, A.H.; Dea, I.C.; Rees, D.A. The fine structures of carob and guar galactomannans. Carbohydr. Res. 1985, 139, 237–260.
- Comfort, D.A.; Chhabra, S.R.; Conners, S.B.; Chou, C.-J.; Epting, K.L.; Johnson, M.R.; Jones, K.L.; Sehgal, A.C.; Kelly, R.M. Strategic biocatalysis with hyperthermophilic enzymes. Green Chem. 2004, 6, 459–465.
- McCutchen, C.M.; Duffaud, G.D.; Leduc, P.; Petersen, A.R.H.; Tayal, A.; Khan, S.A.; Kelly, R.M. Characterization of extremely thermostable enzymatic breakers (α-1,6-galactosidase and β-1,4-mannanase) from the hyperthermophilic bacterium Thermotoga neapolitana 5068 for hydrolysis of guar gum. Biotechnol. Bioeng. 1996, 52, 332–339.
- Tjon-Joe-Pin, R.M. Enzyme breaker for galactomannan based fracturing fluids. U.S. Patent 5,806,597 A, 13 April 1993.
- Liebl, W.; Wagner, B.; Schellhase, J. Properties of an α-galactosidase, and structure of its gene galA, within an α-and β-galactoside utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst. Appl. Microbiol. 1998, 21, 1–11.
- Politz, O.; Krah, M.; Thomsen, K.K.; Borriss, R. A highly thermostable endo-(1,4)- β-mannanase from the marine bacterium Rhodothermus marinus. Appl. Microbiol. Biotechnol. 2000, 53, 715–721.
- You, J.; Liu, J.-F.; Yang, S.-Z.; Mu, B.-Z. Low-temperature-active and salt-tolerant β-mannanase from a newly isolated Enterobacter sp. strain N18. J. Biosci. Bioeng. 2016, 121, 140–146.
- Fridjonsson, O.; Watzlawick, H.; Gehmeiler, A.; Rohrhirsch, T.; Mattes, R. Cloning of the gene encoding a novel thermostable alpha-galactosidase from Thermus brockianus ITI360. Appl. Environ. Microbiol. 1999, 64, 3955–3963.
- Ghosh, B.; Abdelrahim, M.; Belhaj., H. Delayed breaker systems to remove residual polymer damage in hydraulically fractured reservoirs. ACS Omega 2021, 6, 31646–31657.
- Cobianco, S.; Albonico, P.; Battistel, E.; Bianchi, D.; Fornaroli, M. Thermophilic Enzymes for Filtercake Removal at High Temperature. SPE Eur. Form. Damage Conf. 2007, 1–9.
- Brannon, H.D.; Tjon-Joe-Pin, R.M.; Carman, P.S.; Wood., W.D. Enzyme breaker technologies: A decade of improved well stimulation. SPE Annual Technical Conference and Exhibition, Denver, CO, USA, 5–8 October; 2003.
- Zhou, J.; Legemah, M.; Beall, B.; Sun, H.; Qu, Q. Alternative Polysaccharide Fracturing Fluids for Harsh Reservoir Conditions. SPE Unconv. Resour. Conf. Exhib. Asia Pac. 2013, 1–8.
- Aubert, J.P.; Béguin, P. The biological degradation of cellulose. FEMS Microbiol. Rev. 1994, 13, 25–58.
- Sieger, C.; Kroon, A.; Batelaan, J.; van Ginkel, C. Biodegradation of carboxymethyl celluloses by Agrobacterium CM-1. Carbohydr. Polym. 1995, 27, 137–143.
- Lavanya, C.; Kulkarni, P.; Dixit, P.K.; Raavi, L.N.V. Krishna, Sources of cellulose and their applications-A review. Int. J. Drug Formul. Res. 2011, 2, 19–38.
- Ryu, D.D.; Mandels, M. Cellulases: Biosynthesis and applications. Enzym. Microb. Technol. 1980, 2, 91–102.
- Bae, J.; Morisaka, H.; Kuroda, K.; Ueda, M. Cellulosome complexes: Natural biocatalysts as arming microcompartments of enzymes. J. Mol. Microbiol. Biotechnol. 2013, 23, 370–378.
- Poulsen, H.V.; Willink, F.W.; Ingvorsen, K. Aerobic and anaerobic cellulase production by Cellulomonas uda. Arch. Microbiol. 2016, 198, 725–735.
- Dobrev, G.T.; Zhekova, B.Y. Biosynthesis, purification and characterization of endoglucanase from a xylanase producing strain Aspergillus niger B03. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2012, 43, 70–77.
- Saraihom, S.; Kobayashi, D.Y.; Lotrakul, P.; Prasongsuk, S.; Eveleigh, D.E.; Punnapayak, H. First report of a tropical Lysobacter enzymogenes producing bifunctional endoglucanase activity towards carboxymethylcellulose and chitosan. Ann. Microbiol. 2016, 66, 907–919.
- Azizov, E.; Quintero, H.J.; Saxton, K.; Sessarego, S. Carboxymethylcellulose a Cost Effective Alternative to Guar, CMHPG and Surfactant-Based Fluid Systems. SPECSUR Unconv. Resour. Conf. 2015, 1–31.
- Tjon-Joe-Pin, R.M.; Rickards, A.R. Enzyme complex used for breaking crosslinked cellulose based blocking gels at low to moderate temperatures. U.S. Patent 5,224,544 A, 7 July 1993.
- Freeman, M.; Norman, M.; Ballard, D.; Jiang, P.; Symes, K.; Mistry, K. Method and composition for the triggered release of polymer-degrading agents for oil field use. U.S. Patent 20,050,130,845 A1, 16 June 2005.
- Gupta, D.V.S.; Prasek, B.B. Method for fracturing subterranean formations using controlled release breakers and compositions useful therein. U.S. Patent 5,437,331 A, August 15, 1995.
- Trabelsi, S.; Kakadjian, S. Comparative Study Between Guar and Carboxymethylcellulose Used as Gelling Systems in Hydraulic Fracturing Application. SPE Prod. Oper. Symp. 2013, 1–20.
- Scheffer, G.; Berdugo-Clavijo, C.; Sen, A.; Gieg, L.M. Enzyme biotechnology development for treating polymers in hydraulic fracturing operations. Microb. Biotechnol. 2021, 14, 953–966.
- Scheffer, G.; Rachel, N.M.; Ng, K.K.; Sen, A.; Gieg, L.M. Preparation and identification of carboxymethyl cellulose-degrading enzyme candidates for oilfield applications. J. Biotechnol. 2022, 347, 18–25.
- Hong, J.; Ye, Y.; Wang, Y.; Zhang, Y. Bioseparation of recombinant cellulose-binding module-proteins by affinity adsorption on an ultra-high-capacity cellulosic adsorbent. Anal. Chim. Acta 2008, 621, 193–199.
- Han, H.; Ling, Z.; Khan, A.; Virk, A.; Kulshrestha, S.; Li, X. Improvements of thermophilic enzymes: From genetic modifications to applications. Bioresour. Technol. 2019, 279, 350–361.
- Xiong, B.; Loss, R.D.; Shields, D.; Pawlik, T.; Hochreiter, R.; Zydney, A.L.; Kumar, M. Polyacrylamide degradation and its implications in environmental systems. Npj Clean Water 2018, 1, 17.
- Sorbie, K.S. Polymer-Improved Oil Recovery; CRC Press: Boca Raton, FL, USA, 2000.
- Zhao, L.; Song, T.; Han, D.; Bao, M.; Lu, J. Hydrolyzed polyacrylamide biotransformation in an up-flow anaerobic sludge blanket reactor system: Key enzymes, functional microorganisms, and biodegradation mechanisms. Bioprocess Biosyst. Eng. 2019, 42, 941–951.
- Caulfield, M.J.; Qiao, G.G.; Solomon, D.H. Some aspects of the properties and degradation of polyacrylamides. Chem. Rev. 2002, 102, 3067–3084.
- Kay-Shoemake, J.L.; Watwood, M.E.; Lentz, R.D.; Sojka, R.E. Polyacrylamide as an organic nitrogen source for soil microorganisms with potential effects on inorganic soil nitrogen in agricultural soil. Soil Biol. Biochem. 1998, 30, 1045–1052.
- Ma, L.; Hu, T.; Liu, Y.; Liu, J.; Wang, Y.; Zhou, J.; Chen, M.; Yang, B.; Li, L. Combination of biochar and immobilized bacteria accelerates polyacrylamide biodegradation in soil by both bio-augmentation and bio-stimulation strategies. J. Hazard. Mater. 2021, 405, 1–12.
- Song, T.; Li, S.; Lu, Y.; Yan, D.; Sun, P.; Bao, M.; Li, Y. Biodegradation of hydrolyzed polyacrylamide by a Bacillus megaterium strain SZK-5: Functional enzymes and antioxidant defense mechanism. Chemosphere 2019, 231, 183–193.
- Bao, M.; Chen, Q.; Li, Y.; Jiang, G. Biodegradation of partially hydrolyzed polyacrylamide by bacteria isolated from production water after polymer flooding in an oil field. J. Hazard. Mater. 2010, 184, 105–110.
- Grula, M.M.; Huang, M.-L.; Sewell, G. Interactions of certain polyacrylamides with soil bacteria. Soil Sci. 1994, 158, 291–300.
- Haveroen, M.E.; MacKinnon, M.D.; Fedorak, P.M. Polyacrylamide added as a nitrogen source stimulates methanogenesis in consortia from various wastewaters. Water Res. 2005, 39, 3333–3341.
- Hu, H.; Liu, J.-F.; Li, C.-Y.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Anaerobic biodegradation of partially hydrolyzed polyacrylamide in long-term methanogenic enrichment cultures from production water of oil reservoirs. Biodegradation 2018, 29, 233–243.
- Wen, Q.; Chen, Z.; Zhao, Y.; Zhang, H.; Feng, Y. Biodegradation of polyacrylamide by bacteria isolated from activated sludge and oil-contaminated soil. J. Hazard. Mater. 2010, 175, 955–959.
- Kay-Shoemake, J.L.; Watwood, M.E.; Sojka, R.E.; Lentz, R.D. Polyacrylamide as a substrate for microbial amidase in culture and soil. Soil Biol. Biochem. 1998, 30, 1647–1654.
- Liu, L.; Wang, Z.; Lin, K.; Cai, W. Microbial degradation of polyacrylamide by aerobic granules. Environ. Technol. 2012, 33, 1049–1054.
- Yu, F.; Fu, R.; Xie, Y.; Chen, W. Isolation and characterization of polyacrylamide-degrading bacteria from dewatered sludge. Int. J. Environ. Res. Public Health 2015, 12, 4214–4230.
- Ma, F.; Wei, L.; Wang, L.; Chang, C.C. Isolation and identification of the sulphate-reducing bacteria strain H1 and its function for hydrolysed polyacrylamide degradation. Int. J. Biotechnol. 2008, 10, 55–63.
- Nakamiya, K.; Kinoshita, S. Isolation of polyacrylamide-degrading bacteria. J. Ferment. Bioeng. 1995, 80, 418–420.
- Sang, G.; Pi, Y.; Bao, M.; Li, Y.; Lu, J. Biodegradation for hydrolyzed polyacrylamide in the anaerobic baffled reactor combined aeration tank. Ecol. Eng. 2015, 84, 121–127.
- Dai, X.; Luo, F.; Zhang, D.; Dai, L.; Chen, Y.; Dong, B. Waste-activated sludge fermentation for in biological polyacrylamide removal. Sci. Rep. 2015, 5, 1–13.
- Dai, X.; Luo, F.; Yi, J.; He, Q.; Dong, B. Biodegradation of polyacrylamide by anaerobic digestion under mesophilic condition and its performance in actual dewatered sludge system. Bioresour. Technol. 2013, 153, 55–61.
- Yan, M.; Zhao, L.; Bao, M.; Lu, J. Hydrolyzed polyacrylamide biodegradation and mechanism in sequencing batch biofilm reactor. Bioresour. Technol. 2016, 207, 315–321.
- Song, T.; Li, S.; Ding, W.; Li, H.; Bao, M.; Li, Y. Biodegradation of hydrolyzed polyacrylamide by the combined expanded granular sludge bed reactor-aerobic biofilm reactor biosystem and key microorganisms involved in this bioprocess. Bioresour. Technol. 2018, 263, 153–162.
- Berdugo-Clavijo, C.; Sen, A.; Seyyedi, M.; Quintero, H.; O’Neil, B.; Gieg, L.M. High temperature utilization of PAM and HPAM by microbial communities enriched from oilfield produced water and activated sludge. AMB Exp. 2019, 9, 1–10.
- Gupta, D.V.S. Method of using asparaginase as a polyacrylamide enzyme breaker. U.S. Patent 9,090,815 B2, 28 July 2015.
- Ramsden, D.; Fielding, S.; Atkinson, N.; Boota, M. The degradation of polyacrylamide in aqueous solution induced by chemically generated hydroxyl radicals—Part III: Xanthine/xanthine oxidase. Polym. Degrad. Stab. 1987, 17, 49–55.
- Nakamiya, K.; Ooi, T.; Kinoshita, S. Degradation of synthetic water-soluble polymers by hydroquinone peroxidase. J. Ferment. Bioeng. 1997, 84, 213–218.
- Gilbert, W.J.R.; Johnson, S.J.; Tsau, J.-S.; Liang, J.-T.; Scurto, A.M. Enzymatic degradation of polyacrylamide in aqueous solution with peroxidase and H2O2. J. Appl. Polym. Sci. 2016, 134, 1–10.
This entry is offline, you can click here to edit this entry!
