Since the first reported case of COVID-19 in 2019 in China and the official declaration from the World Health Organization in March 2021 as a pandemic, fast and accurate diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has played a major role worldwide. For this reason, various methods have been developed, comprising reverse transcriptase-polymerase chain reaction (RT-PCR), immunoassays, clustered regularly interspaced short palindromic repeats (CRISPR), reverse transcription loop-mediated isothermal amplification (RT-LAMP), and bio(mimetic)sensors. Among the developed methods, RT-PCR is so far the gold standard. IUPAC defines the term biomimetic as “Refers a laboratory procedure designed to imitate a natural chemical process. Also refers to a compound that mimics a biological material in structure or function“. The lotus effect at a water-repelling surface is the best-known example of biomimetic systems. One important motivation for the development and application of biomimetic recognition elements is their potentially higher stability and lower price as compared with biomolecules.
- molecularly imprinted polymers
- biomimetic sensors
- SARS-CoV-2
1. Molecularly Imprinted Polymers
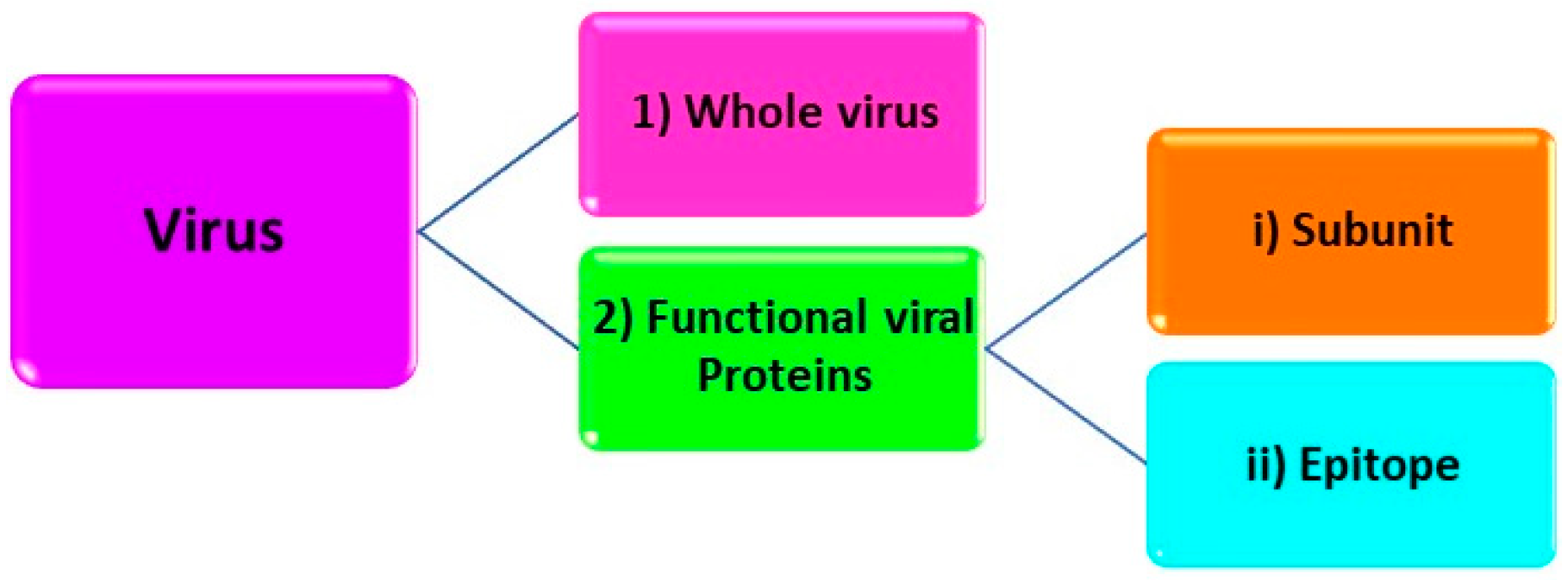
1.1. Structural Levels Target Analytes
1.2. Steps of MIP Preparation
2. MIP-Based Biomimetic Sensors for SARS-CoV-2 Detection
2.1. Electrochemical Detection of SARS-CoV-2
2.2. Optical Detection of SARS-CoV-2
2.3. Commercial MIP for SARS-CoV-2
This entry is adapted from the peer-reviewed paper 10.3390/biomimetics7020058
References
- Yarman, A.; Scheller, F.W. How reliable is the electrochemical readout of MIP sensors? Sensors 2020, 20, 2677.
- Ertürk, G.; Uzun, L.; Tümer, M.A.; Say, R.; Denizli, A. Fab fragments imprinted SPR biosensor for real-time human immunoglobulin G detection. Biosens. Bioelectron. 2011, 28, 97–104.
- Jetzschmann, K.J.; Yarman, A.; Rustam, L.; Kielb, P.; Urlacher, V.B.; Fischer, A.; Weidinger, I.M.; Wollenberger, U.; Scheller, F.W. Molecular LEGO by domain-imprinting of cytochrome P450 BM3. Colloids Surf. B Biointerfaces 2018, 164, 240–246.
- Caserta, G.; Zhang, X.; Yarman, A.; Supala, E.; Wollenberger, U.; Gyurcsányi, R.E.; Zebger, I.; Scheller, F.W. Insights in electrosynthesis, target binding, and stability of peptide-imprinted polymer nanofilms. Electrochim. Acta 2021, 381, 138236.
- Dickert, F.L.; Hayden, O.; Lieberzeit, P.; Haderspoeck, C.; Bindeus, R.; Palfinger, C.; Wirl, B. Nano- and micro-structuring of sensor materials—From molecule to cell detection. Synth. Met. 2003, 138, 65–69.
- Hayden, O.; Lieberzeit, P.A.; Blaas, D.; Dickert, F.L. Artificial antibodies for bioanalyte detection—Sensing viruses and proteins. Adv. Funct. Mater. 2006, 16, 1269–1278.
- Klangprapan, S.; Choke-arpornchai, B.; Lieberzeit, P.A.; Choowongkomon, K. Sensing the classical swine fever virus with molecularly imprinted polymer on quartz crystal microbalance. Heliyon 2020, 6, e04137.
- Altintas, Z.; Gittens, M.; Guerreiro, A.; Thompson, K.A.; Walker, J.; Piletsky, S.; Tothill, I.E. Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers. Anal. Chem. 2015, 87, 6801–6807.
- Gast, M.; Sobek, H.; Mizaikoff, B. Advances in imprinting strategies for selective virus recognition a review. TrAC—Trends Anal. Chem. 2019, 114, 218–232.
- Yarman, A.; Kurbanoglu, S.; Zebger, I.; Scheller, F.W. Simple and robust: The claims of protein sensing by molecularly imprinted polymers. Sens. Actuators B Chem. 2021, 330, 129369.
- Navakul, K.; Sangma, C.; Yenchitsomanus, P.-t.; Chunta, S.; Lieberzeit, P.A. Enhancing sensitivity of QCM for dengue type 1 virus detection using graphene-based polymer composites. Anal. Bioanal. Chem. 2021, 413, 6191–6198.
- Altintas, Z. Advanced Imprinted Materials for Virus Monitoring. In Advanced Molecularly Imprinting Materials; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 389–411.
- Dickert, F.L.; Hayden, O.; Bindeus, R.; Mann, K.-J.; Blaas, D.; Waigmann, E. Bioimprinted QCM sensors for virus detection—Screening of plant sap. Anal. Bioanal. Chem. 2004, 378, 1929–1934.
- Yang, B.; Gong, H.; Chen, C.; Chen, X.; Cai, C. A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus. Biosens. Bioelectron. 2017, 87, 679–685.
- Ramanaviciene, A.; Ramanavicius, A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004, 20, 1076–1082.
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Ramanaviciene, A.; Ciplys, E.; Juozapaitis, M.; Slibinskas, R.; Bechelany, M.; Ramanavicius, A. Molecularly imprinted polypyrrole based sensor for the detection of SARS-CoV-2 spike glycoprotein. Electrochim. Acta 2022, 403, 139581.
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029.
- Amouzadeh Tabrizi, M.; Fernández-Blázquez, J.P.; Medina, D.M.; Acedo, P. An ultrasensitive molecularly imprinted polymer-based electrochemical sensor for the determination of SARS-CoV-2-RBD by using macroporous gold screen-printed electrode. Biosens. Bioelectron. 2022, 196, 113729.
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sens. Actuators B Chem. 2022, 353, 131160.
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288.
- Singh, M.; Gupta, N.; Raghuwanshi, R. Epitope Imprinting Approach to Monitor Diseases. J. Mol. Genet. Med. 2017, 11, 2–7.
- Iskierko, Z.; Sharma, P.S.; Noworyta, K.R.; Borowicz, P.; Cieplak, M.; Kutner, W.; Bossi, A.M. Selective PQQPFPQQ Gluten Epitope Chemical Sensor with a Molecularly Imprinted Polymer Recognition Unit and an Extended-Gate Field-Effect Transistor Transduction Unit. Anal. Chem. 2019, 91, 4537–4543.
- Pasquardini, L.; Bossi, A.M. Molecularly imprinted polymers by epitope imprinting: A journey from molecular interactions to the available bioinformatics resources to scout for epitope templates. Anal. Bioanal. Chem. 2021, 413, 6101–6115.
- Cenci, L.; Guella, G.; Andreetto, E.; Ambrosi, E.; Anesi, A.; Bossi, A.M. Guided folding takes a start from the molecular imprinting of structured epitopes. Nanoscale 2016, 8, 15665–15670.
- Yang, K.; Li, S.; Liu, L.; Chen, Y.; Zhou, W.; Pei, J.; Liang, Z.; Zhang, L.; Zhang, Y. Epitope Imprinting Technology: Progress, Applications, and Perspectives toward Artificial Antibodies. Adv. Mater. 2019, 31, 1902048.
- Fresco-Cala, B.; Rajpal, S.; Rudolf, T.; Keitel, B.; Groß, R.; Münch, J.; Batista, A.D.; Mizaikoff, B. Development and characterization of magnetic SARS-CoV-2 peptide-imprinted polymers. Nanomaterials 2021, 11, 2985.
- Fresco-Cala, B.; Mizaikoff, B. Surrogate Imprinting Strategies: Molecular Imprints via Fragments and Dummies. ACS Appl. Polym. Mater. 2020, 2, 3714–3741.
- Teixeira, S.P.B.; Reis, R.L.; Peppas, N.A.; Gomes, M.E.; Domingues, R.M.A. Epitope-imprinted polymers: Design principles of synthetic binding partners for natural biomacromolecules. Sci. Adv. 2021, 7, eabi9884.
- Dietl, S.; Sobek, H.; Mizaikoff, B. Epitope-imprinted polymers for biomacromolecules: Recent strategies, future challenges and selected applications. TrAC—Trends Anal. Chem. 2021, 143, 116414.
- Rachkov, A.; Minoura, N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim. Biophys. Acta—Protein Struct. Mol. Enzymol. 2001, 1544, 255–266.
- Rachkov, A.; Minoura, N. Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J. Chromatogr. A 2000, 889, 111–118.
- Nishino, H.; Huang, C.S.; Shea, K.J. Selective protein capture by epitope imprinting. Angew. Chem.—Int. Ed. 2006, 45, 2393–2396.
- Dechtrirat, D.; Jetzschmann, K.J.; Stöcklein, W.F.M.; Scheller, F.W.; Gajovic-Eichelmann, N. Protein rebinding to a surface-confined imprint. Adv. Funct. Mater. 2012, 22, 5231–5237.
- Batista, A.D.; Rajpal, S.; Keitel, B.; Dietl, S.; Fresco-Cala, B.; Dinc, M.; Groß, R.; Sobek, H.; Münch, J.; Mizaikoff, B. Plastic Antibodies Mimicking the ACE2 Receptor for Selective Binding of SARS-CoV-2 Spike. Adv. Mater. Interfaces 2022, 9, 2101925.
- Dinc, M.; Esen, C.; Mizaikoff, B. Recent advances on core–shell magnetic molecularly imprinted polymers for biomacromolecules. TrAC—Trends Anal. Chem. 2019, 114, 202–217.
- Bognár, Z.; Supala, E.; Yarman, A.; Zhang, X.; Bier, F.F.; Scheller, F.W.; Gyurcsányi, R.E. Peptide epitope-imprinted polymer microarrays for selective protein recognition. Application for SARS-CoV-2 RBD protein. Chem. Sci. 2022, 13, 1263–1269.
- Altintas, Z.; Takiden, A.; Utesch, T.; Mroginski, M.A.; Schmid, B.; Scheller, F.W.; Süssmuth, R.D. Integrated Approaches Toward High-Affinity Artificial Protein Binders Obtained via Computationally Simulated Epitopes for Protein Recognition. Adv. Funct. Mater. 2019, 29, 1807332.
- Zhang, X.; Caserta, G.; Yarman, A.; Supala, E.; Waffo, A.F.T.; Wollenberger, U.; Gyurcsányi, R.E.; Zebger, I.; Scheller, F.W. “Out of Pocket” Protein Binding—A Dilemma of Epitope Imprinted Polymers Revealed for Human Hemoglobin. Chemosensors 2021, 9, 128.
- Zhang, X.; Yarman, A.; Erdossy, J.; Katz, S.; Zebger, I.; Jetzschmann, K.J.; Altintas, Z.; Wollenberger, U.; Gyurcsányi, R.E.; Scheller, F.W. Electrosynthesized MIPs for transferrin: Plastibodies or nano-filters? Biosens. Bioelectron. 2018, 105, 29–35.
- Wulff, G.; Sarhan, A. Macromolecular Colloquium. Angew. Chem. Int. Ed. Engl. 1972, 11, 334–342.
- Shea, K.J.; Thompson, E. Template synthesis of macromolecules. Selective functionalization of an organic polymer. J. Org. Chem. 1978, 43, 4253–4255.
- Meier, F.; Mizaikoff, B. Molecularly Imprinted Polymers as Artificial Receptors. In Artificial Receptors for Chemical Sensors; Wiley: Weinheim, Germany, 2010; pp. 391–437.
- Yarman, A.; Turner, A.P.F.; Scheller, F.W. Electropolymers for (Nano-)Imprinted Biomimetic Biosensors; Woodhead Publishing Limited: Sawston, UK, 2014; ISBN 9780857096609.
- Arshady, R.; Mosbach, K. Synthesis of Substrate-selective Polymers by Host-Guest Polymerizatioa. Die Makromol. Chem. Macromol. Chem. Phys. 1981, 692, 687–692.
- Whitcombe, M.J.; Rodriguez, M.E.; Villar, P.; Vulfson, E.N. A New Method for the Introduction of Recognition Site Functionality into Polymers Prepared by Molecular Imprinting: Synthesis and Characterization of Polymeric Receptors for Cholesterol. J. Am. Chem. Soc. 1995, 117, 7105–7111.
- D’Aurelio, R.; Tothill, I.E.; Salbini, M.; Calò, F.; Mazzotta, E.; Malitesta, C.; Chianella, I. A comparison of EIS and QCM NanoMIP-based sensors for morphine. Nanomaterials 2021, 11, 3360.
- Viveiros, R.; Rebocho, S.; Casimiro, T. Green strategies for molecularly imprinted polymer development. Polymers 2018, 10, 306.
- Mirsky, V.M.; Hirsch, T.; Piletsky, S.A.; Wolfbeis, O.S. A spreader-bar approach to molecular architecture: Formation of stable artificial chemoreceptors. Angew. Chem.—Int. Ed. 1999, 38, 1108–1110.
- Zhang, X.; Du, X.; Huang, X.; Lv, Z. Creating protein-imprinted self-assembled monolayers with multiple binding sites and biocompatible imprinted cavities. J. Am. Chem. Soc. 2013, 135, 9248–9251.
- Lee, W.-I.; Subramanian, A.; Mueller, S.; Levon, K.; Nam, C.-Y.; Rafailovich, M.H. Potentiometric Biosensors Based on Molecular-Imprinted Self-Assembled Monolayer Films for Rapid Detection of Influenza A Virus and SARS-CoV-2 Spike Protein. ACS Appl. Nano Mater. 2022, 5, 5045–5055.
- Chunta, S.; Suedee, R.; Boonsriwong, W.; Lieberzeit, P.A. Biomimetic sensors targeting oxidized-low-density lipoprotein with molecularly imprinted polymers. Anal. Chim. Acta 2020, 1116, 27–35.
- Basan, H.; Dinc, M.; Mizaikoff, B. Inhibitor-assisted synthesis of molecularly imprinted microbeads for protein recognition. Anal. Methods 2018, 10, 997–1005.
- Tian, X.; Song, H.; Wang, Y.; Tian, X.; Tang, Y.; Gao, R.; Zhang, C. Hydrophilic magnetic molecularly imprinted nanobeads for efficient enrichment and high performance liquid chromatographic detection of 17beta-estradiol in environmental water samples. Talanta 2020, 220, 121367.
- Garcia Lopez, J.; Piletska, E.V.; Whitcombe, M.J.; Czulak, J.; Piletsky, S.A. Application of molecularly imprinted polymer nanoparticles for degradation of the bacterial autoinducer: N-hexanoyl homoserine lactone. Chem. Commun. 2019, 55, 2664–2667.
- Huang, W.; Zhou, X.; Luan, Y.; Cao, Y.; Wang, N.; Lu, Y.; Liu, T.; Xu, W. A sensitive electrochemical sensor modified with multi-walled carbon nanotubes doped molecularly imprinted silica nanospheres for detecting chlorpyrifos. J. Sep. Sci. 2020, 43, 954–961.
- Lian, W.; Liu, S.; Yu, J.; Xing, X.; Li, J.; Cui, M.; Huang, J. Electrochemical sensor based on gold nanoparticles fabricated molecularly imprinted polymer film at chitosan-platinum nanoparticles/graphene-gold nanoparticles double nanocomposites modified electrode for detection of erythromycin. Biosens. Bioelectron. 2012, 38, 163–169.
- Lorenzo, R.A.; Carro, A.M.; Alvarez-Lorenzo, C.; Concheiro, A. To remove or not to remove? The challenge of extracting the template to make the cavities available in molecularly imprinted polymers (MIPs). Int. J. Mol. Sci. 2011, 12, 4327–4347.
- Pirzada, M.; Altintas, Z. Template Removal in Molecular Imprinting: Principles, Strategies, and Challenges. In Molecular Imprinting for Nanosensors and Other Sensing Applications; Elsevier: Amsterdam, The Netherlands, 2021; pp. 367–406.
- Yoshimi, Y.; Ohdaira, R.; Iiyama, C.; Sakai, K. ‘Gate effect’ of thin layer of molecularly-imprinted poly(methacrylic acid-co-ethyleneglycol dimethacrylate). Sens. Actuators B Chem. 2001, 73, 49–53.
- Sharma, P.S.; Garcia-Cruz, A.; Cieplak, M.; Noworyta, K.R.; Kutner, W. ‘Gate effect’ in molecularly imprinted polymers: The current state of understanding. Curr. Opin. Electrochem. 2019, 16, 50–56.
- Hussein, H.A.; Kandeil, A.; Gomaa, M.; Mohamed El Nashar, R.; El-Sherbiny, I.M.; Hassan, R.Y.A. SARS-CoV-2-Impedimetric Biosensor: Virus-Imprinted Chips for Early and Rapid Diagnosis. ACS Sens. 2021, 6, 4098–4107.
- Kalecki, J.; Iskierko, Z.; Cieplak, M.; Sharma, P.S. Oriented Immobilization of Protein Templates: A New Trend in Surface Imprinting. ACS Sens. 2020, 5, 3710–3720.
- Cennamo, N.; Agostino, G.D.; Perri, C.; Arcadio, F.; Chiaretti, G.; Parisio, E.M.; Camarlinghi, G.; Vettori, C.; Di Marzo, F.; Cennamo, R.; et al. Proof of Concept for a Quick and Highly Sensitive On-Site Detection of SARS-CoV-2 by Plasmonic Optical Fibers and Molecularly Imprinted Polymers. Sensors 2021, 21, 1681.
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly Imprinted Polymers in Electrochemical and Optical Sensors. Trends Biotechnol. 2019, 37, 294–309.
- Garcia-Cruz, A.; Haq, I.; Di Masi, S.; Trivedi, S.; Alanazi, K.; Piletska, E.; Mujahid, A.; Piletsky, S.A. Design and fabrication of a smart sensor using in silico epitope mapping and electro- responsive imprinted polymer nanoparticles for determination of insulin levels in human plasma. Biosens. Bioelectron. 2020, 169, 112536.
- Garcia-Cruz, A.; Ahmad, O.S.; Alanazi, K.; Piletska, E.; Piletsky, S.A. Generic sensor platform based on electro-responsive molecularly imprinted polymer nanoparticles (e-NanoMIPs). Microsyst. Nanoeng. 2020, 6, 83.
