Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
A disintegrin and metalloproteinase (ADAM) proteins are proteolytic enzymes that are responsible for destroying the extracellular matrix, but they also have adhesive properties. Recent investigations have demonstrated that the expression of several ADAMs is upregulated in gastrointestinal (GI) tumour cells and have linked the secretion of these proteins to pathogenesis of GI malignancies.
- ADAM
- biomarker
- gastrointestinal tumours
1. Gastrointestinal Cancers—General Characteristics
According to estimates from the American Cancer Society, in 2018 there were approximately 1.7 million new cancer cases and 610,000 cancer deaths [1]. Gastrointestinal (GI) cancers are defined as a group of malignancies that includes cancers of the liver, oesophagus, gallbladder, pancreas, stomach, large and small intestine and anus [2][3][4]. Neoplasms originating from the GI tract, including colorectal cancer (CRC) and gastric cancer (GC), are among the five most common malignancies in both men and women worldwide. It is estimated that CRC is one of the most frequently diagnosed cancers and the second most common cause of cancer-related deaths worldwide. Gastric cancer is the fourth most common cancer and accounts for 8.8% of all cancer-related deaths [2][3][4].
The incidence of GI cancers shows significant geographical variation. CRC incidence is higher in Western Europe and North America, while the incidence of GC and liver cancer (LC) is elevated in Asia and Africa. The main risk factors for GI cancers include tobacco and alcohol intake, genetic factors, viruses such as Papillomaviruses, Epstein–Barr virus (EBV) or hepatitis B and C. Other risk indicators that may cause tumour development are bacterial infections and microbiome imbalance, Helicobacter pylori infection, as well as an unhealthy diet and obesity. It was suggested that Fusobacterium nucleatum plays a role in CRC development via promotion of tumour progression include generating a proinflammatory tumour-promoting microenvironment [5][6][7]. Moreover, the study of Yoshimura revealed that Helicobacter pylori stimulated temporal changes in the levels of proteolytic enzymes such as ADAM10 and ADAM17 transcripts in gastric epithelial cells, while chronic infection with Helicobacter pylori may result in persistent mucosal increases in members of the ADAM family [8]. Some epidemiological studies have shown an increased risk of GI cancers in overweight and obese individuals, while substantial evidence has linked reduced physical activity with an increased risk of colon cancer. Moreover, it has been proven that a high salt intake is associated with enhanced prevalence of GC, whereas a diet high in red and processed meats has been linked to an increased risk of GC, EC, PC and CRC [2][3][4][5][6][7][8][9]. Furthermore, changes in lifestyle, the growing population and environmental factors as well as advances in medicine may also affect the epidemiology of GI cancers [2].
Clinical symptoms of GI cancers depend on the type of malignancy and tumour stage as well as the development of systemic symptoms such as early satiety nausea, anorexia, changes in the sense of smell, stress or dysgeusia [10]. It has been revealed that fatigue is a major sign followed by pain, anxiety, poor well-being, sleep disturbances, poor appetite, depression, drowsiness, dyspnoea and nausea [10][11]. Moreover, in EC, GC and PC, signs of disease may occur early. However, symptoms are directly related to the cancer or release of inflammatory cytokines [10][11]. Neoplasms of the alimentary tract are characterized by rapid progression and a very unfavourable prognosis. The diagnostic process of GI cancers includes endoscopic evaluation as an important tool in diagnosis and staging. Diagnosis is confirmed with an upper gastrointestinal endoscopy and biopsy. Other imaging tests useful in diagnosis of GI tumours include computed tomography (CT), positron emission tomography–CT (PET–CT) and endoscopic ultrasound (EUS) [12]. It is also recommended that apart from imaging tests, diagnosis of GI tumours should include laboratory tests. Measurements of the well-investigated classical tumour markers for GI malignancies such as carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA 19.9), cancer antigen 50 (CA 50) or cancer antigen 72.4 (CA 72.4) are not useful in the early detection of these malignancies due to their low diagnostic sensitivity and specificity. A number of candidates for novel biochemical markers for GI malignancies such as matrix metalloproteinases and their tissue inhibitors, cytokines and chemokines as well as specific proteins such as C-reactive protein or interleukin-6 have been evaluated by scientists in the last decade. However, no studies have confirmed their significance in early diagnosis of GI malignancies [13][14][15][16][17][18][19][20][21][22][23]. Therefore, future research should focus on the search for new, non-invasive and easily accessible biomarkers characterized by high diagnostic sensitivity and specificity, which would be useful in the early detection and tumour staging as well as improve treatment implementation.
Remodelling of the extracellular matrix (ECM) plays an important role in tumour progression, including growth, proliferation and angiogenesis. Moreover, many authors link these proteases to tumour invasiveness, particularly metastasis [13]. Therefore, in the previous studies researchers assessed the usefulness of selected MMPs and their tissue inhibitors in the diagnosis and progression of GI malignancies such as CRC, PC, EC and GC. In addition, selected MMPs, such as MMP-14, are able to regulate a variety of signalling pathways and cell functions, including apoptosis. It has been proven that the proteolytic activity of MMPs is physiologically inhibited by tissue inhibitors of metalloproteinases (TIMPs). TIMP-2 regulates several cell functions including migration, proliferation and apoptosis through MMP-dependent and -independent mechanisms, e.g., via inhibition of FGF-2-induced endothelial cell proliferation, suppression of the mitogenic activity of epidermal growth factor (EGF) or inhibition of angiogenic factor-induced endothelial cell proliferation and angiogenesis [24].
2. A Disintegrin and Metalloproteinase (ADAM)—General Information
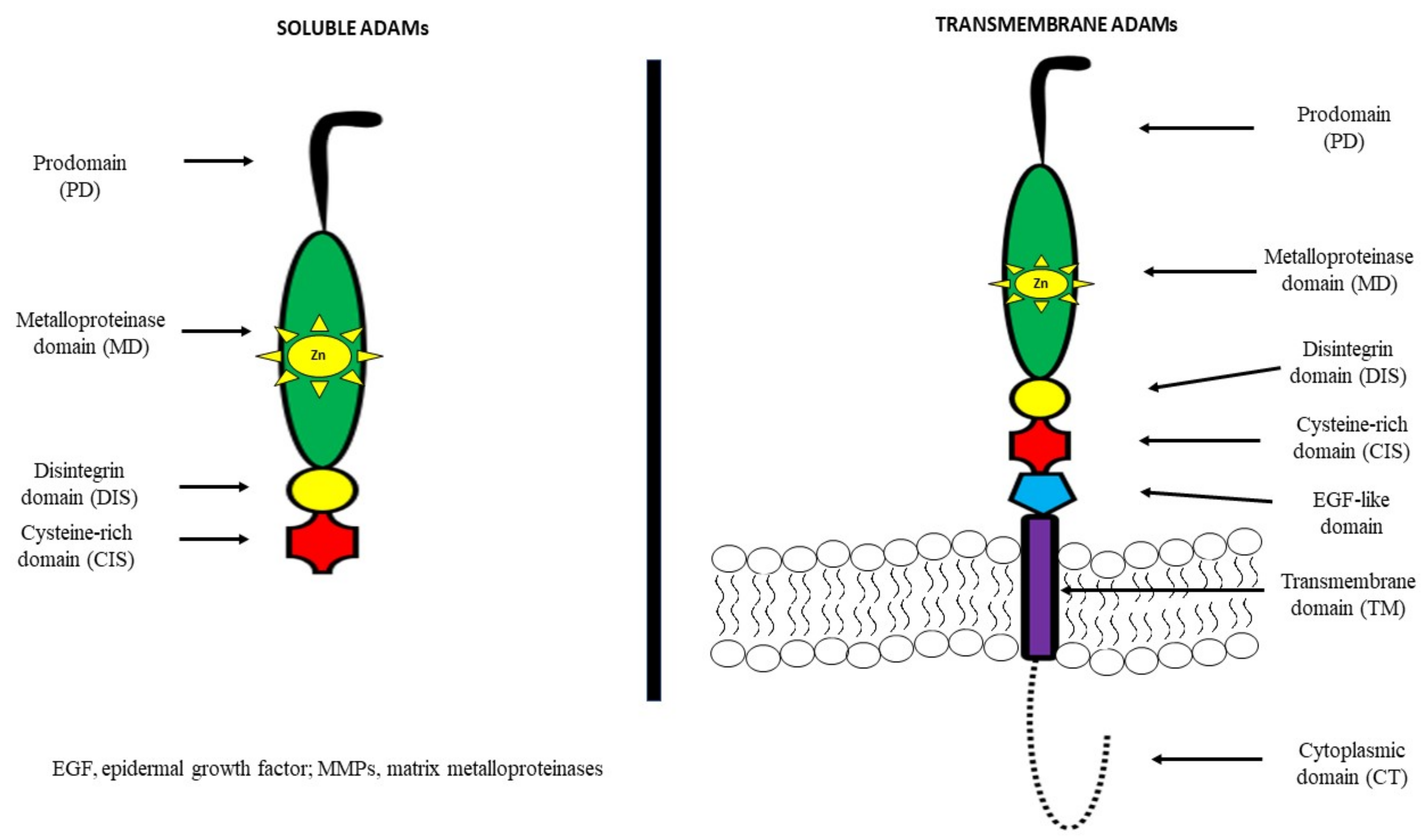
A disintegrin and metalloproteinases (ADAMs) belong to the family of zinc-dependent proteases, such as metalloproteinases, which consist of 21 members, 13 of which have proteolytic activity [25][26]. They are also known as metalloproteinase, disintegrin, cysteine-rich (MDC) proteins. ADAMs regulate the shedding of membrane-bound proteins, cytokines, growth factors as well as ligands and receptors. It has been demonstrated that the structure of most of these proteins consists of a prodomain, a metalloprotease region, a disintegrin domain for adhesion, a cysteine-rich region, epidermal-growth-factor (EGF) repeats, a transmembrane module as well as a cytoplasmic tail [27][28].
It has been observed that among other cell-surface proteins, ADAMs are unique because of both adhesive and proteolytic activities. Moreover, it has been indicated that EGF repeats and the cysteine-rich region mediate cell fusion or the interaction of these proteins with other molecules [29][30][31][32]. ADAMs are mostly transmembrane proteins, but selected ADAMs, such as ADAM11, 12, 17 and 28 may generate a soluble, secreted protein. About 50% of the ADAM family consists of a metalloproteinase domain with a catalytic site consensus sequence that allows for protein–protein interactions [27][28]. The structures of both transmembrane and soluble ADAMs are presented in Figure 1 [30][31][32].
Several ADAM genes initiate more than one protein due to differential splicing of mRNA, a post-transcriptional modification in which a single gene can code for multiple proteins. This promotes the synthesis of a secreted ADAM structure, in addition to membrane-anchored forms, or variation in the length of the cytoplasmic tail of ADAM proteins. According to differences in the active site sequence of the metalloproteinase domain, 60% of the members are non-proteolytic ADAM molecules. By contrast, active sites in the metalloproteinase domain of the proteinase-type ADAM molecules (ADAM8, 9, 10, 12, 15, 17, 19–21, 28, 30 and 33) contain a common HEXGHXXGXXHD sequence with a ‘Met-turn’ which is also present in the catalytic metalloproteinase domain of MMP members [33]. ADAM10 and ADAM17 have a similar structure and are characterized by a membrane-proximal domain in the extracellular region in place of the EGF-like, cysteine-rich domain that provides substrate recognition [34][35]. A distinct subfamily of ADAMs is a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), which consists of a pro-domain and a disintegrin, metalloproteinase, and cysteine-rich domain. In addition, ADAMTS have a specific thrombospondin motif instead of a transmembrane domain [27][28][29].
Growing evidence indicates that ADAM proteases are expressed in an inactive form. It has been proven that the activity of ADAMs and ADAMTS is regulated via endogenous inhibitors called tissue inhibitors of metalloproteinases (TIMPs) as well as proprotein convertases. In addition, they might also be controlled by protein kinase C activators, G-protein coupled receptor agonists or Ca2+ ionophores. Various ADAMs are usually kept in an inactive state because of the interaction of a cysteine residue at the propeptide domain with zinc in the metalloproteinase module. Therefore, ADAMs and MMPs are activated by the cysteine switch mechanism that disrupts the cysteine–zinc interaction to expose the catalytic site. It has been observed that levels of these proteins are regulated at the transcriptional level. However, current knowledge concerning the role of physiological inhibitors of ADAMs is still limited. It has been proven that only TIMP3 inhibits the crystal structure of the protease domain of human ADAM17, while ADAM10 might be inhibited by TIMP1 and 3, and by hydroxamates [35]. TIMP3 may be an inhibitor of various ADAMs as well as ADAMTS members [27][28][29]. Increasing numbers of reports have demonstrated that GI cancer cells might be transformed by aberrant and uncontrolled mechanisms that may produce alternative splicing. It was found that the APC gene, aberrant splice skipping of exon 4, as well as Ron gen, skipping of exon 11 are involved in colon cancer progression. An alternative 5’ splice site in BCL-X, involved in apoptosis, is overexpressed in hepatocellular carcinoma, similar to the CDH17 gene (exclusion of exon 13) that regulates incidence of tumour recurrence. In addition, the TACC1 gene has splicing variants associated with GC. These findings suggest the expression of aberrant and abnormal splice variants in GI cancer development [36][37][38][39].
ADAMs, similarly to MMPs, possess various physiological functions and the ability to regulate many processes such as cell migration, proliferation, angiogenesis, apoptosis, wound healing, tissue repair and survival. By way of illustration, ADAM1 and 2 are able to modulate cell adhesion or sperm–egg fusion. In addition, ADAM12 plays a role in myoblast fusion, while ADAM9, 10 and 17 regulate ectodomain shedding of cell-surface proteins [27][28][29]. However, these molecules might also be involved in some pathological conditions such as cardiovascular and malignant diseases, including GI cancers [40][41][42][43].
3. A Disintegrin and Metalloproteinase (ADAM)—Their Role in Tumour Development
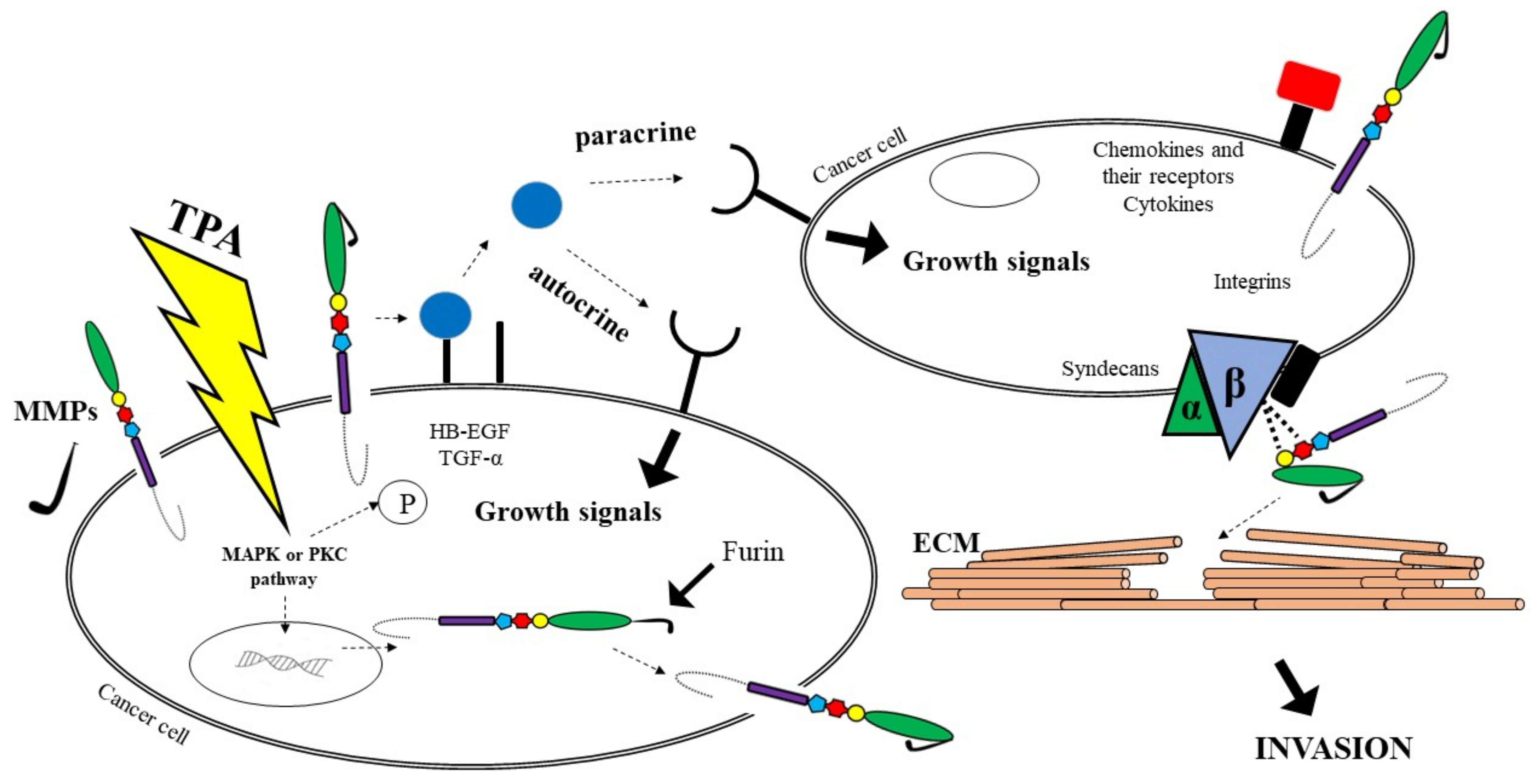
A growing body of evidence suggests that cancer growth is not only driven by tumour cell-intrinsic mechanisms, but might be dependent on paracrine signals, such as growth factors or cytokines, produced by the tumour microenvironment (TME). These molecules are synthesized as trans-membrane proteins and must be released by limited proteolysis defined as ectodomain shedding. It has been proven that ADAMs are major mediators of ectodomain shedding and thus are able to initiate paracrine signal transduction [25]. Changes in the activation process of paracrine signal transduction is a crucial step in the development of GI malignancies. Therefore, ADAM proteases play an important role in inflammation as well as pathogenesis of several tumours because of their ability to interact with a variety of substrates [25].
The multiple functional roles of ADAMs prove their involvement in a variety of normal and pathophysiological conditions, including cancer progression [32][41][42][44]. Characteristics of selected ADAMs [32] and their significance in tumour biology are presented in Table 1.
Table 1. Characteristics of selected ADAMs [32].
| ADAMs | Other Name | Involvement in Cancer Biology | Inhibitors |
|---|---|---|---|
| ADAM8 | MS2 (CD156) | Promotion of migration | - |
| ADAM9 | MDC9, MCMP, Meltrin-γ | Promotion of cell adhesion and invasion, binding to integrins (α6β4 and α2β1) | - |
| ADAM10 | MDAM, Kuzbanian | Type I membrane glycoprotein L1 shedding, promotion of cell growth and migration | TIMP1 TIMP3 |
| ADAM12 | Meltrin-α, MCMP, MLTN, MLTNA | HB-EGF (heparin-binding epidermal growth factor) shedding, promotion of cell growth | TIMP3 |
| ADAM15 | Metargidin, MDC15, AD56, CR II-7 | Promotion of cell growth | No data |
| ADAM17 | TACE, cSVP | TGF-β (transforming growth factor) shedding, promotion of cell growth | TIMP2 TIMP3 |
| ADAM19 | Meltrin-β, FKSG34 | No data | - |
| ADAM28 | e-MDC II, MDC-Lm, MDC-Ls | IGFBP-3 (insulin-like growth factor binding protein-3) cleavage, promotion of cell growth | TIMP3 TIMP4 |
| ADAMTS1 | C3-C5, METH1, KIAA1346 | HB-EGF (heparin-binding epidermal growth factor) and AR shedding, promotion of cell growth, survival and invasion | No data |
| ADAMTS4 | KIAA0688, aggrecanase-1, ADMP-1 | No data | TIMP3 |
| ADAMTS5 | ADAMTS11, aggrecanase-2, ADMP-2 | Brevican cleavage, promotion of invasion | TIMP3 |
It has been reported that ADAM8 is involved in tumour cell migration and invasion, ADAM9 plays a role in tumorigenesis, invasion and metastasis through modulation of growth factor activity and integrin function, while overexpression of ADAM10 appears to promote the growth and proliferation of tumour cells. In addition, ADAM12 cleaves various ECM molecules including gelatin, type IV collagen and fibronectin, suggesting a potential role of this enzyme in ECM digestion in cancer invasion and metastasis. Thus, ADAM12 functions as a shedder, adhesion molecule and ECM-degrading proteinase and is involved in cancer progression. ADAM28 may play a key role in cancer cell proliferation and metastasis, whereas ADAM17 is a target of tumorigenesis, but the role of ADAM15 in cancer biology remains to be elucidated [32][44].
It has been established that ADAM12 and ADAM28 regulate the level of free IGF-1 by proteolysis of the IGFBP-3/IGF-1 protein complex [44]. In addition, ADAM17 protein is responsible for the activation of TNFα, initiating the signalling pathway associated with the EGF receptor for which it is a ligand, leading to tumour cell proliferation [45]. Adamalysines might be involved in the pathogenesis of gastric cancer via the EGFR signalling pathway and the TGF-α/Smad pathway [46]. In vitro assays indicate that ADAM8 overexpression promotes cell growth and increases migration and invasion abilities by decreasing the p-p38/p-extracellular regulated protein kinase (p-ERK) ratio. Several studies suggest that there are five most common pathways of ADAMs involvement in cancer biology. It has been reported that proADAM might be activated via furin or MMPs. As furin activates MMP activity, cancer cells have the potential to become metastatic. Another pathway leads to growth factors such as TGFα shedding, which may change signals on the cancer cell’s surface. Soluble growth factors activate EGFR on cells, causing the enhanced cells proliferation via autocrine and paracrine manners. A third route involves the participation of ADAMs as adhesion molecules with integrins on cells, which may facilitate the digestion of the substrates of the ECM. Fourth, cell proliferation signals can be regulated indirectly by ADAMs via integrins; thus these molecules provide traction to migrating cells through the ECM using integrins. In addition, ADAMs are able to stimulate cancer development and metastasis via the interaction with other molecules, including cytokines and their receptors, that are also associated with cancer progression. ADAMs, similar to MMPs, are able to cleave ECM molecules. As a consequence, it allows neoplastic cells to adhere to new locations, which is responsible for, e.g., cancer metastasis [44]. All the pathways are presented in Figure 2.
Recent studies indicate the importance of the ADAM family in tumour formation, migration, proliferation and development [47][48]. There is increasing evidence that several ADAMs are differentially expressed in tumours. Some studies have confirmed the role of ADAMs in the biology of malignant cells, including breast [49][50], renal [51] and small cell lung [52] cancer as well as GI malignancies such as gastric [46][47][48][49][50][51][52][53][54][55][56][57][58], colorectal [59][60][61][62] and pancreatic [63][64][65] cancer and hepatocellular carcinoma [66][67][68][69].
This entry is adapted from the peer-reviewed paper 10.3390/cancers14092307
References
- Cancer Facts & Figures 2018. Atlanta: American Cancer Society. 2018. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf (accessed on 26 April 2022).
- Dizdar, Ö.; Kılıçkap, S. Global Epidemiology of Gastrointestinal Cancers; Yalcin, S., Philip, P.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–12.
- Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/cancer-facts-and-figures-2019.pdf (accessed on 26 April 2022).
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33.
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, J.; Tommasino, M.; et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1381–1390.
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980.
- Abed, J.; Maalouf, N.; Manson, A.L.; Earl, A.M.; Parhi, L.; Emgård, J.E.M.; Klutstein, M.; Tayeb, S.; Almogy, G.; Atlan, K.A.; et al. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front. Cell. Infect. Microbiol. 2020, 10, 400.
- Yoshimura, T.; Tomita, T.; Dixon, M.F.; Axon, A.T.R.; Robinson, P.A.; Crabtree, J.E. ADAMs (a disintegrin and metalloproteinase) messenger RNA expression in Helicobacter pylori-infected, normal, and neoplastic gastric mucosa. J. Infect. Dis. 2002, 185, 332–340.
- D’Elia, L.; Galletti, F.; Strazzullo, P. Dietary salt intake and risk of gastric cancer. Cancer Treat. Res. 2014, 159, 83–95.
- Yavuzsen, T.; Kazaz, N.; Tanriverdi, Ö.; Akman, T.; Davis, M.P. Symptom Management in Gastrointestinal Cancers; Yalcin, S., Philip, P.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 669–685.
- Hui, D.; Shamieh, O.; Paiva, C.E.; Perex-Cruz, P.E.; Kwon, J.H.; Muckaden, M.A.; Park, M.; Yennu, S.; Kang, J.K.; Bruera, E. Minimal clinically important differences in the Edmonton Symptom Assessment Scale in cancer patients: A prospective, multicenter study. Cancer 2015, 121, 3027–3035.
- Karaosmanoglu, A.D.; Onur, M.R.; Arellano, R.S. Imaging in Gastrointestinal Cancers; Yalcin, S., Philip, P.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 445–464.
- McCawley, L.J.; Matrisian, L.M. Matrix metalloproteinases: Multifunctional contributors to tumor progression. Mol. Med. Today 2000, 6, 149–156.
- Łukaszewicz-Zając, M.; Gryko, M.; Pączek, S.; Szmitkowski, M.; Kędra, B.; Mroczko, B. Matrix metalloproteinase 2 (MMP-2) and its tissue inhibitor 2 (TIMP-2) in pancreatic cancer (PC). Oncotarget 2019, 10, 395–403.
- Mroczko, B.; Kozłowski, M.; Groblewska, M.; Łukaszewicz, M.; Nikliński, J.; Jelski, W.; Laudański, J.; Chyczewski, L.; Szmitkowski, M. The diagnostic value of the measurement of matrix metalloproteinase 9 (MMP-9), squamous cell cancer antigen (SCC) and carcinoembryonic antigen (CEA) in the sera of esophageal cancer patients. Clin. Chim. Acta 2008, 389, 61–66.
- Mroczko, B.; Łukaszewicz-Zając, M.; Wereszczyńska-Siemiątkowska, U.; Groblewska, M.; Gryko, M.; Kędra, B.; Jurkowska, G.; Szmitkowski, M. Clinical significance of the measurements of serum matrix metalloproteinase-9 and its inhibitor (tissue inhibitor of metalloproteinase-1) in patients with pancreatic cancer. Metalloproteinase-9 as an independent prognostic factor. Pancreas 2009, 38, 613–618.
- Mroczko, B.; Łukaszewicz-Zając, M.; Guzińska-Ustymowicz, K.; Gryko, M.; Czyżewska, J.; Kemona, A.; Kędra, B.; Szmitkowski, M. Expression of matrix metalloproteinase-9 in the neoplastic and interstitial inflammatory infiltrate cells in gastric cancer. Folia Histochem. Cytobiol. 2009, 47, 491–496.
- Mroczko, B.; Groblewska, M.; Łukaszewicz-Zając, M.; Bandurski, R.; Kędra, B.; Szmitkowski, M. Pre-treatment serum and plasma levels of matrix metalloproteinase 9 (MMP-9) and tissue inhibitor of matrix metalloproteinases 1 (TIMP-1) in gastric cancer patients. Clin. Chem. Lab. Med. 2009, 47, 1133–1139.
- Mroczko, B.; Łukaszewicz-Zając, M.; Gryko, M.; Kędra, B.; Szmitkowski, M. Clinical significance of serum levels of matrix metalloproteinase 2 (MMP-2) and its tissue inhibitor (TIMP-2) in gastric cancer. Folia Histochem. Cytobiol. 2011, 49, 125–131.
- Melton, S.D.; Genta, R.M.; Souza, R.F. Biomarkers and Molecular Diagnostic Tests in Gastrointestinal Tract and Pancreatic Neoplasms. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 620–628.
- Łukaszewicz-Zając, M.; Mroczko, B. Circulating Biomarkers of Colorectal Cancer (CRC)—Their Utility in Diagnosis and Prognosis. J. Clin. Med. 2021, 10, 2391.
- Łukaszewicz-Zając, M.; Mroczko, B.; Gryko, M.; Kędra, B.; Szmitkowski, M. Comparison between clinical significance of serum proinflammatory proteins (IL-6 and CRP) and classic tumor markers (CEA and CA 19-9) in gastric cancer. Clin. Exp. Med. 2011, 11, 89–96.
- Pawluczuk, E.; Łukaszewicz-Zając, M.; Gryko, M.; Kulczyńska-Przybik, A.; Mroczko, B. Serum CXCL8 and Its Specific Receptor (CXCR2) in Gastric Cancer. Cancers 2021, 13, 5186.
- Valacca, C.; Tassone, E.; Mignatti, P. TIMP-2 Interaction with MT1-MMP Activates the AKT Pathway and Protects Tumor Cells from Apoptosis. PLoS ONE 2015, 10, e0136797.
- Schumacher, N.; Rose-John, S.; Schmidt-Arras, D. ADAM-Mediated Signalling Pathways in Gastrointestinal Cancer Formation. Int. J. Mol. Sci. 2020, 21, 5133.
- Edwards, D.R.; Handsley, M.M.; Pennington, C.J. The ADAM metalloproteinases. Mol. Asp. Med. 2008, 29, 258–289.
- Mentlein, R.; Hattermann, K.; Held-Feindt, J. Lost in disruption: Role of proteases in glioma invasion and progression. Biochim. Biophys. Acta 2012, 1825, 178–185.
- Englund, A.T.; Geffner, M.E.; Nagel, R.A.; Lippe, B.M.; Braunstein, G.D. Pediatric germ cell and human chorionic gonadotropin producing tumors. Clinical and laboratory features. Am. J. Dis. Child. 1991, 145, 1294–1297.
- Uhm, J.H.; Dooley, N.P.; Villemure, J.G.; Yong, V.W. Glioma invasion in vitro: Regulation by matrix metalloprotease-2 and protein kinase C. Clin. Exp. Metastasis 1996, 14, 421–433.
- Haoyuan, M.A.; Yanshu, L.I. Structure, regulatory factors and cancer-related physiological effects of ADAM9. Cell Adhes. Migr. 2020, 14, 165–181.
- Giebeler, N.; Zigrino, P.A. Disintegrin and Metalloprotease (ADAM): Historical Overview of Their Functions. Toxins 2016, 8, 122.
- Mochizuki, S.; Okada, Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007, 98, 621–628.
- Lorenzen, I.; Lokau, J.; Düsterhöft, S.; Trad, A.; Garbers, C.; Scheller, J.; Rose-John, S.; Grötzinger, J. The membrane-proximal domain of A Disintegrin and Metalloprotease 17 (ADAM17) is responsible for recognition of the interleukin-6 receptor and interleukin-1 receptor II. FEBS Lett. 2012, 586, 1093–1100.
- Düsterhöft, S.; Michalek, M.; Kordowski, F.; Oldefest, M.; Sommer, A.; Röseler, J.; Reiss, K.; Grötzinger, J.; Lorenzen, I. Extracellular Juxtamembrane Segment of ADAM17 Interacts with Membranes and Is Essential for Its Shedding Activity. Biochemistry (Moscow) 2015, 54, 5791–5800.
- Schlondorff, J.; Blobel, C.P. Metalloprotease-disintegrins: Modular proteins capable of promoting cell–cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 1999, 112, 3603–3617.
- Ghigna, C.; Giordano, S.; Shen, H.; Benvenuto, F.; Castiglioni, F.; Comoglio, P.M.; Green, M.R.; Riva, G.; Biamonti, S. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 2005, 20, 881–890.
- Takehara, T.; Liu, X.; Fujimoto, J.; Friedman, S.L.; Takahashi, H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology 2001, 34, 55–61.
- Line, A.; Slucka, Z.; Stengrevics, A.; Li, G.; Rees, R.C. Altered splicing pattern of TACC1 mRNA in gastric cancer. Cancer Genet. Cytogenet. 2002, 139, 78–83.
- Kim, Y.-J.; Kim, H.-S. Alternative Splicing and Its Impact as a Cancer Diagnostic Marker. Genom. Inform. 2012, 10, 74–80.
- Chute, M.; Jana, S.; Kassiri, Z. Disintegrin and metalloproteinases (ADAMs and ADAM-TSs), the emerging family of proteases in heart physiology and pathology. Curr. Opin. Physiol. 2018, 1, 34–45.
- Duffy, M.J.; Mullooly, M.; O’Donovan, N.; Sukor, S.; Crown, J.; Pierce, A.; McGowan, P.M. The ADAMs family of proteases: New biomarkers and therapeutic targets for cancer? Clin. Proteom. 2011, 8, 9.
- Duffy, M.J.; McKiernan, E.; O’Donovan, N.; McGowan, P.M. Role of ADAMs in cancer formation and progression. Clin. Cancer Res. 2009, 15, 1140–1144.
- Walkiewicz, K.; Kozieł, P.; Bednarczyk, M.; Błażelonis, A.; Mazurek, U.; Muc-Wierzgoń, M. Expression of Migration-Related Genes in Human Colorectal Cancer and Activity of a Disintegrin and Metalloproteinase 17. Biomed. Res. Int. 2016, 2016, 8208904.
- Mochizuki, S.; Shimoda, M.; Shiomi, T.; Fujii, Y.; Okada, Y. ADAM28 is activated by MMP-7 (matrilysin-1) and cleaves insulin-like growth factor binding protein-3. Biochem. Biophys. Res. Commun. 2004, 315, 79–84.
- Gao, M.Q.; Kim, B.G.; Kang, S.; Choi, Y.P.; Yoon, J.H.; Cho, N.H. Human breast cancer-associated fibroblasts enhance cancer cell proliferation through increased TGF-α cleavage by ADAM17. Cancer Lett. 2013, 336, 240–246.
- Carl-McGrath, S.; Lendeckel, U.; Ebert, M.; Roessner, A.; Röcken, C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int. J. Oncol. 2005, 26, 17–24.
- Wagstaff, L.; Kelwick, R.; Decock, J.; Edwards, D.R. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front. Biosci. 2011, 16, 1861–1872.
- Herat, L.; Rudnicka, C.; Okada, Y.; Mochizuki, S.; Schlaich, M.; Matthews, V. The Metalloproteinase ADAM28 Promotes Metabolic Dysfunction in Mice. Int. J. Mol. Sci. 2017, 18, 884.
- Roy, R.; Moses, M.A. ADAM12 induces estrogen-independence in breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 731–741.
- Roy, R.; Rodig, S.; Bielenberg, D.; Zurakowski, D.; Moses, M.A. ADAM12 transmembrane and secreted isoforms promote breast tumor growth: A distinct role for ADAM12-S protein in tumor metastasis. J. Biol. Chem. 2011, 286, 20758–20768.
- Fritzsche, F.R.; Wassermann, K.; Jung, M.; Tölle, A.; Kristiansen, I.; Lein, M.; Johannsen, M.; Dietel, M.; Jung, K.; Kristiansen, G. ADAM9 is highly expressed in renal cell cancer and is associated with tumour progression. BMC Cancer 2008, 26, 179.
- Shao, S.; Li, Z.; Gao, W.; Yu, G.; Liu, D.; Pan, F. ADAM-12 as a diagnostic marker for the proliferation, migration and invasion in patients with small cell lung cancer. PLoS ONE 2014, 9, e85936.
- Ni, P.; Yu, M.; Zhang, R.; He, M.; Wang, H.; Chen, S.; Duan, G. Prognostic Significance of ADAM17 for Gastric Cancer Survival: A Meta-Analysis. Medicina 2020, 56, 322.
- Huang, J.; Bai, Y.; Huo, L.; Xiao, J.; Fan, X.; Yang, Z.; Chen, H.; Yang, Z. Upregulation of a disintegrin and metalloprotease 8 is associated with progression and prognosis of patients with gastric cancer. Transl. Res. 2015, 166, 602–613.
- Kim, J.M.; Jeung, H.C.; Rha, S.Y.; Yu, E.J.; Kim, T.S.; Shin, Y.K.; Zhang, X.; Park, K.H.; Park, S.W.; Chung, H.C.; et al. The effect of disintegrin-metalloproteinase ADAM9 in gastric cancer progression. Mol. Cancer Ther. 2014, 13, 3074–3085.
- Wang, Y.Y.; Ye, Z.Y.; Li, L.; Zhao, Z.S.; Shao, Q.S.; Tao, H.Q. ADAM 10 is associated with gastric cancer progression and prognosis of patients. J. Surg. Oncol. 2011, 103, 116–123.
- Xu, M.; Zhou, H.; Zhang, C.; He, J.; Wei, H.; Zhou, M.; Lu, Y.; Sun, Y.; Ding, J.W.; Zeng, J.; et al. ADAM17 promotes epithelial-mesenchymal transition via TGF-β/Smad pathway in gastric carcinoma cells. Int. J. Oncol. 2016, 49, 2520–2528.
- Yin, Q.; Gu, J.; Qi, Y.; Lu, Y.; Yang, L.; Liu, J.; Liang, X. ADAM28 from both endothelium and gastric cancer cleaves von Willebrand Factor to eliminate von Willebrand Factor-induced apoptosis of gastric cancer cells. Eur. J. Pharmacol. 2021, 898, 173994.
- Dosch, J.; Ziemke, E.; Wan, S.; Luker, K.; Welling, T.; Hardiman, K.; Fearon, E.; Thomas, S.; Flynn, M.; Rios-Doria, J.; et al. Targeting ADAM17 inhibits human colorectal adenocarcinoma progression and tumor-initiating cell frequency. Oncotarget 2017, 8, 65090–65099.
- Yang, Z.; Bai, Y.; Huo, L.; Chen, H.; Huang, J.; Li, J.; Fan, X.; Yang, Z.; Wang, L.; Wang, J. Expression of A disintegrin and metalloprotease 8 is associated with cell growth and poor survival in colorectal cancer. BMC Cancer 2014, 14, 568.
- Walkiewicz, K.; Strzelczyk, J.; Waniczek, D.; Biernacki, K.; Muc-Wierzgoń, M.; Copija, A.; Nowakowska-Zajdel, E. Adamalysines as Biomarkers and a Potential Target of Therapy in Colorectal Cancer Patients: Preliminary Results. Dis. Markers 2019, 2019, 5035234.
- Toquet, C.; Colson, A.; Jarry, A.; Bezieau, S.; Volteau, C.; Boisseau, P.; Merlin, D.; Laboisse, C.L.; Mosnier, J.F. ADAM15 to α5β1 integrin switch in colon carcinoma cells: A late event in cancer progression associated with tumor dedifferentiation and poor prognosis. Int. J. Cancer 2012, 130, 278–287.
- Yamada, D.; Ohuchida, K.; Mizumoto, K.; Ohhashi, S.; Yu, J.; Egami, T.; Fujita, H.; Nagai, E.; Tanaka, M. Increased expression of ADAM 9 and ADAM 15 mRNA in pancreatic cancer. Anticancer Res. 2007, 27, 793–799.
- Valkovskaya, N.; Kayed, H.; Felix, K.; Hartmann, D.; Giese, N.A.; Osinsky, S.P.; Friess, H.; Kleeff, J. ADAM8 expression is associated with increased invasiveness and reduced patient survival in pancreatic cancer. J. Cell Mol. Med. 2007, 11, 1162–1174.
- Alldinger, I.; Dittert, D.; Peiper, M.; Fusco, A.; Chiappetta, G.; Staub, E.; Lohr, M.; Jesnowski, R.; Baretton, G.; Ockert, D.; et al. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology 2005, 5, 370–379.
- Yuan, S.; Lei, S.; Wu, S. ADAM10 is overexpressed in human hepatocellular carcinoma and contributes to the proliferation, invasion and migration of HepG2 cells. Oncol. Rep. 2013, 30, 1715–1722.
- Shiu, J.S.; Hsieh, M.J.; Chiou, H.L.; Wang, H.L.; Yeh, C.B.; Yang, S.F.; Chou, Y.E. Impact of ADAM10 gene polymorphisms on hepatocellular carcinoma development and clinical characteristics. Int. J. Med. Sci. 2018, 15, 1334–1340.
- Zhang, Y.; Tan, Y.F.; Jiang, C.; Zhang, K.; Zha, T.Z.; Zhang, M. High ADAM8 expression is associated with poor prognosis in patients with hepatocellular carcinoma. Pathol. Oncol. Res. 2013, 19, 79–88.
- Zhang, W.; Liu, S.; Liu, K.; Wang, Y.; Ji, B.; Zhang, X.; Liu, Y. A disintegrin and metalloprotease (ADAM)10 is highly expressed in hepatocellular carcinoma and is associated with tumor progression. J. Int. Med. Res. 2014, 42, 611–618.
This entry is offline, you can click here to edit this entry!