Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
In the era of multi-omic sciences, which continuously provide a more comprehensive overview of human pathogenesis, dogma on singular cause-effect in physio-pathological processes is overcome and system biology approaches have been welcomed to look at pathologies through new perspectives. In this context, extracellular vesicles (EVs) have also been targets of this approach, shedding light on their heterogeneity in terms of content, function, origin and potentiality.
- surfaceomics
- system biology
- EVs origin
1. Proteomics
EVs proteomics explicates its potentiality as it might suggest numerous answers to tricky questions, following well-defined protocols and taking advantage of many different approaches, such as gel-free and gel-based approaches(Figure 1).

Figure 1. EVs proteomic workflow. First step is the sample collection and EVs isolation starting commonly from biological fluids and cell lines. Proteins are extracted and prepared for proteomic analysis. By gel-based approach, a 2DE proteomic profiling and differential analysis is performed, followed by MALDI-TOF/TOF protein identification. By gel-free approach, proteins are digested enzymatically and processed by LC-MS/MS and dedicated software. Identified proteins are then subjected to biological interpretation by bioinformatic enrichment analysis such as network and pathway analyses. Some illustrations were adapted from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License.
As commonly accepted, EVs’ heterogeneity in term of subtypes is far beyond the classical classification by size or biogenesis and the difficulty to discriminate them within a whole biological sample represents a limit in analyzing their protein cargoes, how these change across the subpopulations and how these associate with a pathogenic process [50]. In the recent years, there have been various attempts to comparatively analyze the protein cargo of different EVs populations, often comparing small-EVs (s-EVs) and large-EVs (l-EVs). Although several common proteins are identified, numerous unique proteins are also detected, primarily suggesting potential subtypes markers and distinct and/or common associated molecular pathways [51,52]. Indeed, getting deeper into EVs populations is not just limited to their classification purpose, but it also extends to investigate their biological variability. Haraszti et al. reported a comparative proteomic study of s-EVs and l-EVs isolated from glioblastoma, hepatocellular carcinoma and bone marrow mesenchymal stem cells (MSC). The proteomic data of s-EVs resulted closely related to the donor cells proteome, rather than the l-EVs one, allowing to differentiate cancer cells and MSC [53,54]. As the proteomic profile of EVs is not only related to the vesicular main components, rather it includes an extensive part of proteins strictly related to their progenitor cells, the proteomic approach by two-dimensional electrophoresis (2DE) might retain a valuable potentiality. In our recent paper about the proteomic characterization of EVs from bronchoalveolar lavage fluid (BALF) of idiopathic pulmonary fibrosis (IPF) patients, we compared the 2DE map of EVs isolated from BALF with the 2DE image of the whole biological sample, showing that the obtained protein profiles were different and characteristics [55]. As Figure 2 shows, the 2DE proteomic pattern of IPF BALF-EVs (Figure 2A) is visually unique and dissimilar to that of the whole BALF (Figure 2B), as well as to that of EVs isolated from the BALF of patients with other interstitial lung diseases (ILDs) (Figure 2C). These results first confirm the good isolation of EVs as any or very few contaminants, such as highly abundant protein species normally present in common biological fluids, are visible in the vesicular 2DE maps; secondly, they underline the strong divergent protein content between the whole biological sample and the derived EVs. Remarkably, other proteomic analyses conducted by our research group highlighted how the vesicular protein cargo is distinct from its original sample; Montecchi et al. focused on the differential proteomic analysis by 2DE of murine astrocytes and murine astrocytes-derived EVs [56]. As Figure 3 shows, the 2DE vesicular map (Figure 3A) is clearly different from the proteomic pattern of the cellular culture of origin (Figure 3B) and highly characteristic of its original sample. Furthermore, our research group performed other studies on EVs isolated by different biological samples. In Figure 4A,B, it can be observed the 2DE map of plasma-derived EVs, isolated by a combination of size exclusion chromatography (SEC) and a commercial isolation kit, and of human plasma, respectively. Moreover, Figure 5A,B shows the 2DE proteomic profile of human seminal plasma-derived EVs by commercial isolation kit and of human seminal plasma, respectively. Even through different isolation procedures, the unique proteomic profile of EVs might be observed as clearly distinct from that of the original sample in all of our studies, as well as distinct also between themselves, suggesting 2DE as a good visualization technique of vesicular proteomes and of their correlation to their belonging progenitor sample. Other studies used proteomic approach by 2DE to perform EVs profiling, such as Di Giuseppe et al. in the characterization of two different EVs subpopulations isolated from human glioblastoma stem cell secretome [57], as well as Kitamura et al., who performed a proteomic profiling of exosomes from blood samples of patients affected by Parkinson’s disease [58].
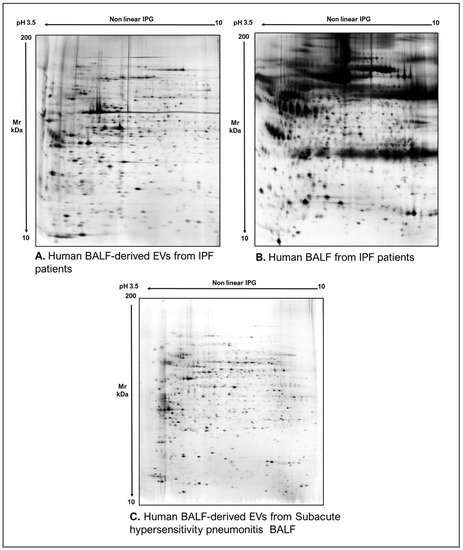
Figure 2. Two-dimensional electrophoresis images indicative of the protein profile of (A) EVs from BAL of IPF patients, (B) BAL of IPF patients and (C) EVs from BAL of subacute hypersensitivity pneumonitis patients.
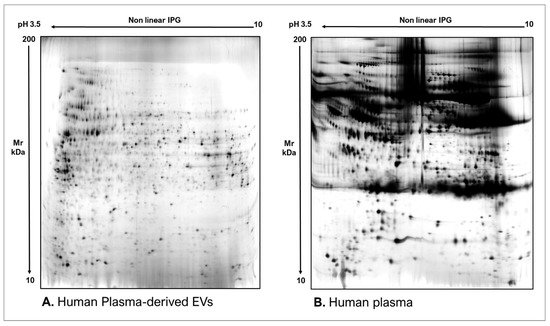
Figure 3. Two-dimensional electrophoresis images indicative of the protein profile of (A) Astrocytes culture-derived EVs and (B) astrocytes cells.
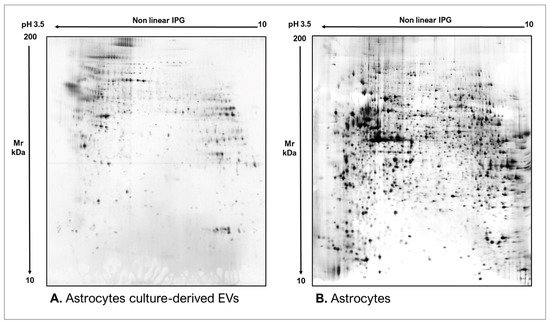
Figure 4. Two-dimensional electrophoresis images indicative of the protein profile of (A) human plasma-derived EVs and (B) human plasma.
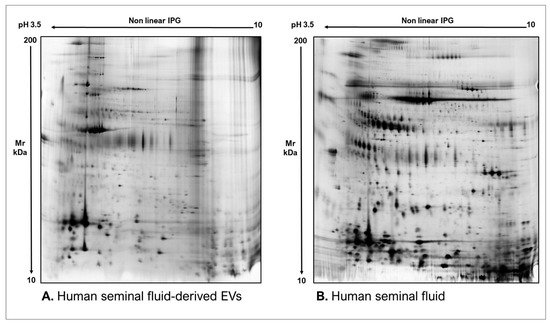
Figure 5. Two-dimensional electrophoresis images indicative of the protein profile of (A) Human seminal fluid-derived EVs and (B) Human seminal fluid.
Of equal importance, mass spectrometry by shotgun approach might provide a wide and highly sensitive range of information on EVs proteomic content, contributing considerably to their characterization and profiling [59,60,61,62,63]. Furthermore, this high-throughput approach has been continuously demonstrating its potentiality in EVs biomarkers discovery, as well as their therapeutical applications [64,65]. For instance, another important application of EVs proteomic profiling is the identification of disease-associated diagnostic and prognostic markers, as EVs proteome is both cell and disease-type dependent. Indeed, EVs are also capable of shaping the microenvironment as active players also in pathological events, such as metastatic promotion and tumor growth promotion [66,67]. The active involvement of EVs in cancer homeostasis and the fact that their composition reflects the contents of cell of origin make them optimal candidates for biomarker profiling studies. Liquid biopsies, indeed, are gaining great popularity given their many advantages in disease diagnosis, and proteomics retains a fundamental role as EVs protein biomarkers allow to overcome limits given by abundant proteins that dominate common biological fluids. For example, Choi et al. reported the identification of CD5L in serum-derived EVs as lung cancer biomarker [68], while Ganig et al. reported QSOX1 as a promising novel EVs biomarker for early diagnosis of colorectal cancer [69]. Other studies focused also on EVs biomarker profiling in other pathologies, such as Nielsen et al. who explored serum-EVs protein biomarkers for Alzheimer’s disease diagnosis [70]. Furthermore, EVs proteome could offer a snapshot of proteomic signatures reflecting on one side diverse stages of diseases progression; for example, recently Pecankova et al. reported a specific clusterin proteoform as a promising biomarker for myelodysplastic syndrome [71], and on the other side mirroring a response to therapy [72,73]. EVs proteomics also contributes considerably to EVs application as drug delivery systems. In fact, therapeutical agents or biological cargo may be encapsulated into EVs and then the vesicles engineered to acquire the requested features for delivery. Moreover, EVs are preferred rather than nanoparticles or liposomes because of their nano size, their ability to cross biological barriers and reduced immunogenicity [74].
2. Transcriptomics
EVs RNA content has been considerably investigated in the past years, reporting the presence of prevalently mRNAs, miRNAs and long non-coding (lnc) RNAs. However, the development of more advanced high-throughput RNA sequencing methods has led to the detection of various other RNA species, such as small nuclear (sn) RNA, small nucleolar (sno) RNA, non-coding (nc) RNA, long intergenic non-coding (linc) RNA, piwi-interacting (pi) RNA, vault RNA, small non-coding (Y)-RNA, small conditional (sc) RNA, circular (circ) RNA, signal recognition particle (SRP)-RNA and 7SK-RNA, as well as fragments originating from rRNA, tRNA, mRNA and lncRNAs [77,78,79]. Given the variety of functions and activities exploited by EVs RNAs, they offer a great chance to determine novel potential biomarkers of screening, diagnosis and prognosis of pathological conditions. For instance, Rhode et al. investigated the expression level of mRNAs encoding for CD44, PTEN and FASN in plasma-derived EVs from patients with gastric cancer, in order to improve differential diagnosis of different subtypes of this tumor [80]. Newman et al. investigated the potentiality of four specific miRNAs in the differential diagnosis of non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) in plasma-derived EVs [81]. Nonetheless, several studies have been demonstrating that miRNAs represent just a minority of the total RNAs contained in EVs, throwing light on the classes of non-coding RNAs [82]. Recently, Iparraguirre et al. investigated the RNA cargo in plasma-derived EVs of multiple sclerosis (MS) patients, reporting a particular enrichment of circRNAs in EVs of MS patients and significant differences in circRNA and linear RNA expression between MS types as well [83]; Magaña et al. evaluated the small ncRNA pattern in EVs released from medulloblastoma (MB) and diffuse infiltrative pontine glioma (DIPG) patient-derived cell lines, identifying novel miRNAs not previously associated to the pathogenesis of the diseases and demonstrating an enrichment of Y-RNAs in the released EVs [84], and Peng et al. evaluated piRNAs in urinary EVs as non-invasive biomarkers for prostate cancer diagnosis [85]. Although scientific community has been giving great attention to the EVs RNA profiling in order to explore their gene-regulatory activities and their potentiality in paving a new direction in therapeutic targeting, the state-of-the-art of EVs transcriptomics is to provide a clear picture of the EVs content, however many improvements are needed to better understand the message mediated by these structures [86]. Summary data are reported in Table A1.
3. Metabolomics and Lipidomics
Despite growing potentiality of metabolomics, EV databases, such as Vesiclepedia, ExoCarta or EVpedia mainly contain protein, mRNA and miRNA entries with less lipid and metabolite data [87,88,89]. Actually, in the last decade metabolomics has been demonstrating its promising contribution to EVs research. Metabolites are represented by any biologically relevant molecule below 2 kDa in size, therefore a wide range of molecules are included, such as steroid hormones, metabolic intermediates, amino acids, nucleotides and different enzymatic cofactors. Its first application is the EVs profiling of the metabolomic content. EVs metabolome, indeed, represents a phenotype screenshot of progenitor’s cellular state, thereby it may provide helpful information on metabolic dysregulations occurring in physio-pathological events [90]. Furthermore, monitoring of changes in EVs metabolome in patient’s biofluids could be extremely useful in providing information on disease progression and response to treatments [91]. Of interest, the major contribution of EVs metabolomics is given by providing encouraging metabolite biomarker candidates. For example, Clos-Garcia et al. reported an elevated level of the steroid hormone dehydroepiandrosterone sulphate (DHEAS) in urinary EVs from prostate cancer patients [92]. In addition, recently Lou et al. identified four metabolites as potential markers for severe acute pancreatitis (SAP) diagnosis, including eicosatrienoic acid (C20:3), thiamine triphosphate, 2-Acetylfuran, and cis-Citral [93]. Beyond clinical applications, EVs metabolome might also provide alternative information on EVs as metabolically active machines. For example, some studies demonstrated the involvement of EVs in the metabolic regulation of the extracellular space, such as in tumor growth and metastatic processes [94]. Remarkably, there is a strong crossover between EV metabolomics and lipidomics, as the size of most relevant lipids identifies them as metabolites [95]. In particular, lipidomics focuses on the characterization and quantification of lipid species as functionally active molecules, as well as on the lipid compositional response to various stimuli. For such reasons, this omic science addresses first a more comprehensive understanding of EVs biogenesis and packaging pathways [96,97]. In particular, numerous EVs lipidomic studies demonstrated that EVs are rich in certain lipids, such as cholesterol, sphingomyelin, phosphatidylserine (PS), phosphatidylcholine (PC), and phosphatidylinositol (PI), suggesting their role as cell-to-cell lipid mediators [98,99]. On the other hand, EVs lipidomes’ characterization is valuable in evaluating lipid dysregulations associated to various diseases. For instance, Su et al. investigated the lipidomic profile of brain-derived EVs (BDEVs) in human frontal cortex to determine a potential altered lipid pattern in Alzheimer’s disease [100]. The other encouraging application of lipidomics is the research of EVs lipid biomarkers.
4. Genomics
Another branch of EVs omic research addresses their genomic cargo, including single strand (ss) DNA, double strand (ds) DNA, mitochondrial (mt) DNA, plasmid DNA, along with nuclear protein histones as well. In addition, DNA could be found either enclosed within EVs or attached to the EVs’ outer surface, or both [18]. The most promising application of EVs genomics is its biomarker use, as EVs DNA reflects the mutational status of parental cells, making the diagnosis and prognosis of different pathological conditions, such as tumors, easier and more accessible. This is supported also by the enhanced availability of EVs DNA in body fluids as protected from degradation by the EVs lipid bilayer [102]. Many examples are given by the extensive research on liquid biopsies of various cancers [103]; for instance, Takur et al. reported the detection of specific colon carcinoma-associated mutations in plasma-derived EV-DNA; Lee et al. tested the sensitivity in detecting epidermal growth factor receptor (EGFR) mutations in EVs-DNA from BALF of patients with advanced non-small cell lung cancer (NSCLC) by targeted next-generation sequencing (NGS) of both tissue DNA and vesicular DNA [104]. Moreover, various studies have evaluated the methylation profile of EVs DNA, proving similarities with the methylation pattern in tissue-derived DNA [102]. For instance, Maire et al. reported that EVs-DNA derived from glioblastoma cells reflects the tumor methylation pattern and mutational profiles of tumor tissue-derived DNA [105]. Of particular interest, EVs genomics shed light also on EVs engineering for therapeutical use, as several studies have been attempting at loading DNA into EVs, still encountering many technical limits [106,107]. Although the biggest goal is to explore the potentiality of these nanostructures in delivering chosen molecules to specific targets, still many questions remain to be elucidated on the natural EVs behaviour itself [108]. Summary data are reported in Table A1.
5. Surfaceomics
Among omic sciences, surfaceomics has been attracting much attention recently. Surfaceome consists of the collection of membrane proteins of which parts are exposed to the extracellular space, for this reason it might be considered responsible for the interaction between the cells and the surrounding environment. Its growing interest is associated to the several functions that the surfaceome carries out and that might be target of biotechnological innovations: first, it regulates the selective permeability of the plasma membrane, which is of particular importance for efficient drug delivery systems to cells; second, the surfaceome is rich in receptors that capture and mediate specific external signals to the internal compartment of the cell, triggering a specific response. Moreover, the surfaceome is also composed of proteins that mediates both a defense and offense against harmful attacks. Due to these features, the surfaceome is a big source of knowledge of physio-pathological processes, as well as of potential pharmaceutical targets [109]. For instance, Rose et al. reported the identification of potential therapeutical targets for the glioblastoma treatment, as they investigated the glioblastoma-associated surfaceome by shotgun proteomics [110]. Another recent innovative surfaceomics application focused on the identification of novel surface antigens in bacterial surfaceome for vaccine development, as reported by Luu et al. [111]. As a result, surfaceomics has found its application also on EVs research. On one side, EVs surfaceomics might provide candidate surface proteins for the enrichment of EVs derived from specific cell types, such as cancer-cell exosomes, enhancing the ability to differentiate EVs produced from diverse tissues and improving EVs-based diagnosis. For example, Castillo et al. carried out the surfaceome profiling of pancreatic ductal adenocarcinoma (PDAC) exosomes by proteomic approach [112]. On the other side, the study of EVs surfaceome might provide powerful tools for diagnostic and therapeutic actions, such as antibody mediated therapy, as well as for drug delivery strategies by engineering EVs with specific target-cell recognition protein on their surface [113,114]. Summary data are reported in Table A1.
This entry is adapted from the peer-reviewed paper 10.3390/proteomes10020012
This entry is offline, you can click here to edit this entry!
