1. Introduction
Low back pain is one of the most frequently observed clinical conditions, and degenerative spine disease seems to be a leading driver of low back pain
[1]. The global prevalence of low back pain increased from 377.5 million in 1990 to 577.0 million in 2017
[2]. The years lived with a disability increased globally from 42.5 million in 1990 to 64.9 million in 2017, representing an increase of 52.7%. Degenerative spinal disease is a common and impairing condition resulting in high socio-economic costs. Direct medical expenses spent on low-back pain doubled to 102 billion USD between 1997 and 2005, and the number of lumbar fusion procedures has quadrupled over the past 20 years, resulting in significantly increased healthcare costs
[2].
Interestingly, the increase in performed surgeries is not directly proportional to improved patient outcomes. Impaired quality of life, persistent pain, and functional problems are reported in up to 40% of patients undergoing low back pain surgery and 20–24% undergoing revision surgeries
[3][4]. Indications influencing the decision as to whether a patient should undergo surgery are not entirely based on guidelines but rather on discussions between the surgeon and patient, as well as the expertise and skills of the surgeon. Furthermore, there are no clear guidelines on surgical techniques for treating degenerative spinal diseases; as such, it remains unclear as to whether one treatment approach might perform better in particular cases than another. Overall, there is a considerable lack of data-driven decision-making in low back pain patients, which is particularly concerning when considering the global burden associated with low back pain.
Medical healthcare is driven by an incredible increase in the amount of data generated through various diagnostic tools and nodes within the healthcare systems. Patient data are the fundaments healthcare providers use to find the best fitting prognosis and diagnosis for each patient. Decisions are based on patterns across these datasets that guide towards the “right” diagnosis. Moreover, prognosis healthcare providers utilize these datasets to justify a specific treatment approach. Therefore, the correct interpretation of these datasets is crucial and directly impacts patient outcomes and the operations of healthcare systems.
Furthermore, improvements in treatment guidelines are mainly based on research that has been performed on such datasets. Researchers using these data might not be aware of the patterns hidden in their collected datasets. The process of finding patterns in large datasets which specifically fall under the category of big-data research is called data mining
[5]. However, oftentimes, clinical researchers might not have profound knowledge in (bio)statistics personally, nor access to biostatisticians, to apply the best available tools to their datasets to extract all relevant pieces of information. Therefore, it is of high relevance that such datasets are made public and anonymized so that data scientists can use them and possibly determine these patterns using modern data-mining technologies.
The term “digital health” stands for the digitalization of healthcare data that was previously only assessed in an unproductive way through paper-based forms. New healthcare applications have become increasingly relevant and available. Such applications can range from mobile health applications, consumer techs, and telehealth for monitoring and guiding patients to precision medicine utilizing patient-specific data in artificial intelligence and bioinformatics models for individualized treatment approaches.
Machine learning combines biostatistics, mathematics, and computer science into one problem-solving pathway. One advantage is its efficiency and effectiveness, as the underlying programming code can be modified to enhance the accuracy of paths that solve a specific task. In this way, it can be more controllable, cost-efficient, and less error-prone than its “human template.” Although the number of publications and citations in artificial-intelligence-related papers on healthcare topics is overwhelming, the technique is still at the beginning of its maturity. The industry highly supports the progress because of the great potential to improve medical research and clinical care, particularly as healthcare providers increasingly establish electronic health records in their institutions.
Predictive analysis with classical statistical techniques, such as regression models, applied on these datasets has been the gold standard to date. One may ask about the advantages of advanced machine learning techniques over simple regression analysis using widely available statistical software for predictive analysis. It is hard to draw a distinct line indicating where basic statistical methods end and machine learning begins. It is often debated whether statistical techniques are somehow also considered as machine learning techniques, as in these cases, computers are using mathematical models to test a specific hypothesis. The primary differentiation might be the purpose of the application. Statistical methods, such as regression models, aim to find associations between independent variables (e.g., age, sex, body mass index) and dependent variables (e.g., patient-related outcome measures). Contrastingly, machine learning models also use statistical methods but aim to learn from training datasets, helping them to make more accurate predictions on the validation dataset so that the model can be reliably used on other independent datasets for predictive analysis. Hence, machine learning could be explained as focusing on predictive results, whereas simple statistical models analyze significant relationships. However, one further differentiation might be interpretability. The more complex a machine learning technique gets, the more accurate it can become at the cost of interpretability. For example, the lasso regression is a machine learning technique using regression analysis for feature selection and prediction. It has the advantage that it is not necessary to find the relevant independent variables first, as is required in linear regression modes. Its application is quite simple, and the interpretability is high. In contrast, deep learning, which is a subgroup of machine learning and will be discussed later, can get very complex but also very accurate; however, this comes at the cost of interpretability. The general principles of machine learning discussed in this research might help to differentiate between the most utilized approaches.
One significant barrier of machine learning applications is that reliable learning processes are very data-hungry. Machine learning is highly dependent on the premise that a large dataset is available. As computers cannot process visual and textual information the way human brains do, the algorithm needs to know what it is predicting or classifying in order to make decisions. When classification tasks need to be solved or specific areas need to be predicted, annotations are necessary. Data annotations help to make the input data understandable for computers. The task of the data scientist is to reliably label data such as text, audio, images, and video so it can be recognized by machine learning models and used to solve prediction and classification tasks. However, this process can be highly time-consuming, which might represent a major flaw in the implementation of machine learning algorithms. Non-accurate labeling will ultimately lead to inaccurate problem-solving. In previous research entitled “Deep Learning: A Critical Appraisal”
[4], Marcus et al. proposed ten concerns associated with machine learning research, and data hungriness was listed as the top factor. He noted that “in problems where data are limited, deep learning often is not an ideal solution”
[6]. Data-hungriness was also considered an unsolved problem in artificial intelligence (AI) research, described in Martin Ford’s book “Architects of Intelligence: the Truth About AI From the People Building It”
[7]. Most of the researchers interviewed in his book encourage the development of more data-efficient algorithms. Four pillars relevant for the implementation and interpretation of machine learning algorithms were described by Cutillo et al. based on discussions at the National Institute of Health (NIH) healthcare workshop in 2019
[8]. These were Trustworthiness, Explainability, Usability, Transparency, and Fairness.
An increase in data efficiency cannot be made feasible only by increasing the number of input samples but also by improving the machine learning architecture itself. One way to do this is to consider that different data types might contribute differently to the problem-solving task and that the connection between data types might also be relevant. Discussing the data dependency of machine learning algorithms and different hybrid models capable of processing different data types is, unfortunately, a research field that has not received the necessary attention yet. In particular, the translation of such hybrid algorithms to a clinical environment with real-world applications has not yet been reviewed.
2. The Need for Structured Decision Making in Spine Surgery
A step towards precision and data-driven spine surgery can be achieved by meeting the significant requirement of developing informative outcomes in assessments. These include regular outcome assessments of patients, preferably utilizing digital app-based assessment forms, and the necessity to implement these outcomes as dependent variables in future risk assessment tools. Notably, the improvement of patient-related outcome measures (PROMs) should be the primary goal of decision-making. The value of such outcome measures is more critical in clinics than surrogate markers such as laboratory markers and classical clinical variables such as revision surgery, readmission, or absence of surgical infections. Previous research has shown that patient-related outcome measures do not necessarily correlate with the factors a surgeon might consider relevant. For example, researchers could show that patient-related outcome measures were more correlated with the length of hospital stay than with postoperative complication rates
[9]. Therefore, patient-related outcome measures should be an integral part of every predictive tool in spine surgery. In spine surgery, commonly utilized patient-related outcome measures include the Oswestry Disability Index, Core Outcome Measure Index (COMI), the eq-5D, SF-36 form, Numeric Rating Scale of pain, and the Visual Analogue Scale of pain, in addition to others
[10]. Notably, Breakwell et al. reported in their publication entitled “Should we all go to the PROM? The first two years of the British Spine Registry” that a significant amount of PROMs forms were entered by the patients themselves
[11]. Hence, an app-based tool transferring the results from the PROMs to a central database could be more time-efficient for spine surgeons. An additional benefit would be that outcomes could be compared considering all contributing institutes, and necessary quality-control steps could be performed in an early phase. This could also be very cost-efficient for healthcare institutes.
The integration of these patient-related outcome measures as dependent variables in clinical decision support tools would allow outcomes to be predicted during prospective follow-ups based on a set of several textual independent variables such as surgical technique, preoperative markers, as well as other data modalities such as imaging. This approach could reliably analyze large volumes of data based on previous data input and suggest next steps for treatment, flag potential problems, and enhance care team efficiency. Furthermore, this PROMs-including approach allows surgeons to discuss the possible outcome with patients and therefore improves the communication with patients. Contrastingly, a communication style in which the surgeon advises against surgery based on his subjective experience might lead to a negative surgeon-patient relationship. Such data-driven support tools might also be better to help surgeons communicate with patients.
3. Database Repositories for Machine Learning Applications in Spine Surgery
Databases are repositing data for future research. They are dedicated to housing data related to scientific research on a platform that can be restricted for access or publicly available. One often-used approach is to limit access to all contributors of the database. In this way, the database integrates a simple reward system: the contribution of data allows contributors to use the gathered data. Databases can collect and store a heterogeneous set of patient data and large datasets that fall under the category of big data. Usually, data in online medical databases are stored anonymously, maintaining that data cannot be linked to the patients’ personal information. In such cases, radiological images can be stored with genetic and clinical data, all having a unique identification number linking the different datatypes of the case. These databases cover a wide range of data, including those related to cancer research, disease burden, nutrition and health, and genetics and the environment. Researchers can apply for access to data based on the scope of the database and the application procedures required to perform relevant medical research.
For machine learning purposes, these data can also be labeled/annotated before being uploaded, allowing for utilization via data scientists. Although impactful machine learning models published to date might deal with a well-annotated dataset, the annotation process requires the necessary infrastructure, expertise, and resources, as it is very time-consuming depending on the number of data points. Considering the complexity of data annotation, crowdsourcing platforms are currently emerging. In this crowdsourcing model, the data are annotated by multiple crowdsourcing workers. One advantage is that the labeling can be checked against the consensus label using statistical parameters such as the inter-annotator agreement. Furthermore, this approach could lead to a more generalizable annotation style within the dataset. Therefore, the model might better predict future datasets coming from other workgroups. Crowdsourcing applications introduced were, for example, applications to database curation, the identification of medical terms in patient-authored texts, and the diagnosis of diseases from medical images
[12][13][14]. Such platforms could also be applied by institutes in spine surgery. Although recent studies have shown that the accuracy performed by crowd workers is mainly similar to the individual annotation considering a given task, crowdsourcing is more resource-oriented and reliable
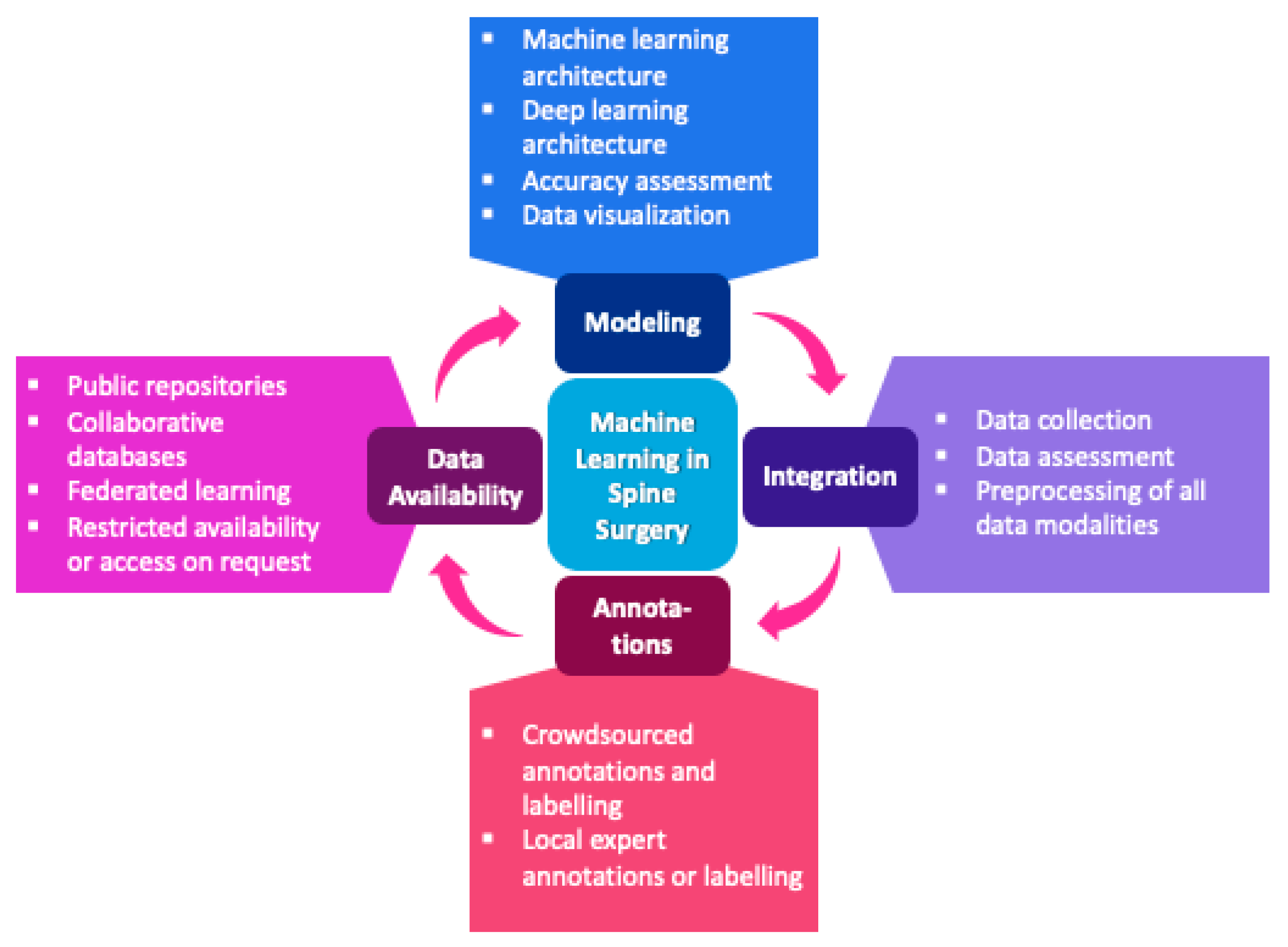
[15][16][17]. The workflow of machine learning applications in spine surgery is shown in
Figure 1.
Figure 1. Workflow of machine learning applications in spine surgery.
Several databases are available that house an impressive number of global biomedical data. Notably, these repositories are regularly updated and extended using new image sets and data types provided by multiple institutions. Thus, they are often used by machine learning research studies, which is essential for progress in the field and exemplary for upcoming databases. For example, the GDC data portal
[18] can provide RNA-sequencing, whole-genome, whole-exome sequencing, targeted sequencing, genotype, tissue and diagnostic slides, and ATAC-seq data. These data types could also be used as an input in hybrid machine learning models along with imaging and clinical data types to solve prediction tasks related to spinal oncology. Access to these platforms can be obtained from researchers but only for subsets of the whole dataset. The general principle of these platforms is that only data that will be used can be extracted. However, all mentioned databases do not contain data labeling and annotations. Considering that there can be vast amounts of data depending on the research question, this might be a significant limitation for using these data for machine learning purposes. Several sources of public databases are accessible by anyone who wants to train and test their machine learning models. One such example is the Kaggle Dataset collection, which contains several algorithms and datasets in spine surgery
[19][20]. These datasets are often used for competitions and training novel machine learning methods to determine whether they outperform existing models. This allows for a peer-review process as the algorithms are publicly available and commented on by other data scientists, validating the algorithm on the dataset provided and external datasets. However, since journal peer-reviewers may not have the resources to retest provided datasets with the algorithm code, often uploaded in GitHub repositories
[21], such open peer-review processes meet crucial research goals, including validity, objectivity, and reliability. Furthermore, provided datasets and codes from the workgroups may not be available after some time. This represents a significant flaw in the assessment and development process of machine learning algorithms for healthcare applications. Publications are not the only relevant output of research; research data should also be considered. This is particularly true when considering that more accurate analysis pathways might not have developed when the study was conducted. This paradigm led to the emergence of data journals, such as Scientific Data from Nature
[22] or GigaScience from Oxford Academic
[23] in which the data can remain available for future analysis and validity assessments.
Notably, in surgical fields, such databases are still scarce. One of the largest and most intuitive databases in orthopedic surgery is the Osteoarthritis Initiative (OAI) database
[24] from the National Institute of Health, which includes ten-year multi-center observational data of knee osteoarthritis cases. It includes DICOM images, clinical data, and laboratory data, and it is one of the few and most extensive repositories in orthopedic surgery capable of integrating multimodal data. Unfortunately, to the best of our knowledge, the only database that seems to include multimodal data in spine surgery is the Austrian Spinal Cord Injury Study
[25]. The database contains longitudinal data on spinal cord injury cases in Austria and includes clinical data with patient-related outcome measures and imaging data. Other databases in spine surgery, which mainly include tabular clinical data, are the American College of Surgeon National Surgical Quality Improvement Project (ASC-NSQIP) database, the National Inpatient Sample (NIS) database, the Medicare and Private Insurance Database, the American Spine Registry, and the British Spine Registry
[11][26][27]. The SORG (“Sorg Orthopaedic Research Group”) has introduced the most recognized and cited predictive machine learning models, which can be accessed for free on their website
[28]. They were already externally validated several times and include mortality prediction algorithms in spinal oncology, PROMs, and postoperative opioid use predictions after spine surgery, as well as discharge disposition for lumbar spinal surgery. Validation and external validation studies are both accessible on the website.
Another emerging field aiming to address the data handling problem in machine learning is privacy-first federated learning
[29]. Federated learning
[30][31] aims to train machine learning algorithms collaboratively without the need to transfer medical datasets. This approach would address the data governance and privacy politics, often limiting the use of medical data depending on the country where the research is conducted. Federated learning was extensively applied in mobile and edge device applications and is currently increasingly applied in healthcare environments
[32][33]. It enables the assessment and development of models collaboratively using peer-review techniques without transferring the medical data out of the institutions where the data were obtained. Instead, machine learning training and testing take place on an institutional level, and only model architecture information and parameters are transferred between the collaborators. Recent studies have shown that machine learning models trained by Federated Learning can achieve similar accuracies to models that were implemented using central databases and are even superior to those processed on a single-institution-level
[34][35]. Successful implementation of Federated Learning approaches could thus hold significant potential for enabling resource-oriented precision healthcare at a large scale, with external validation to overcome selection bias in model parameters and to promote the optimal processing of patients’ data by respecting the necessary governance and privacy policies of the participants
[33]. Nevertheless, this approach still requires essential infrastructure and quality management processes to ensure that the applications perform well and do not impair healthcare processes or violate patient privacy rules.
Despite the advantages of Federated Learning, this method still has some disadvantages. For example, as described above, the integration of medical datasets in public databases could lead to more extensive research, and the investigation would not be limited to the collaborators. Furthermore, successful model training still depends on factors such as data labeling, data quality, bias, and standardization
[36]. These issues would be better targeted when databases are accessible by more researchers and crowdfunding workers dealing with data annotation. This would be the case for both Federated and non-Federated Learning techniques. Appropriate protocols would be required, focusing on well-designed studies, standardized data extractions, standardized labeling and annotation of data, accuracy assessments and quality management, and regularly updated techniques to assess bias or failures. Considering this, Federated Learning would be a feasible approach to overcome data transfer limitations between institutions.
This entry is adapted from the peer-reviewed paper 10.3390/jpm12040509