Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Oxidative stress is the result of an imbalance between the presence of reactive oxygen species (ROS) and the ability of a biological system to detoxify them and their side products. Similarly, nitrosative stress is an imbalance of reactive nitrogen species (RNS). Some of these molecules can play an important role in signalization in both eukaryotes and prokaryotes.
- bacterial envelope
- oxidative stress
- nitrosative stress
1. Oxidative Stress Chemistry
Molecular oxygen (O2) is a small non-polar molecule that diffuses freely across usual biological membranes [1]. Therefore, the bacterial intracellular O2 concentration is similar to their environment. As a result, bacteria either have to avoid oxidative stress by living in anaerobic or microaerobic environments or survive elevated internal oxygen levels. Oxygen is not toxic by itself, as it is practically unreactive with the molecules structuring biological organisms that are lipids, proteins, carbohydrates, and nucleic acids. However, the reduction of O2 can generate various ROS, such as the superoxide anion (O2•−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH•) [2]. In aerobic environments, endogenous ROS may be produced in bacteria via the reaction between O2 and univalent electron donors. These donors can be metal centers, dihydroflavin cofactors, or quinones [2]. The main endogenous source of O2•− and H2O2 is the autoxidation of non-respiratory flavoproteins [3] by electron transfer between O2 and the dihydroflavin of the reduced flavoproteins. This reaction leads to the formation of O2•−, which generally goes through another electron transfer before escaping the active site of the enzyme, generating H2O2. Aside from its conversion into H2O2, O2•− is in equilibrium with the hydroperoxyl radical (HO2•) through the reversible reaction O2•− + H+ ⇋ HO2• [4]. Since the pKa of this reaction is 4.88, it is estimated that the HO2• form represents less than 1% of the total superoxide in the cellular cytoplasm [5]. However, cytoplasmic pH may not be uniform and may be significantly lowered in the proximity of membranes containing negatively charged phospholipids such as cardiolipin, phosphatidylserine, and phosphatidylinositol [6]. Therefore, local pH decrease could allow the formation of significant quantities of HO2• near the IM. Unlike O2•−, HO2• is hydrophobic, allowing it to cross to the IM lipidic core. As a result, HO2• is critical in the process of lipid peroxidation. In the presence of light, photosynthetic organisms also generate the highly reactive singlet oxygen (1O2) through the pigments of their photosystems [7]. This stress can also occur in non-photosynthetic microorganisms through other cellular cofactors such as rhodopsin, quinones, flavins, and porphyrins [7].
2. Nitrosative Stress Chemistry
The range of reactive molecules created by oxidative stress is not limited to ROS. Indeed, RNS production is tightly linked to the presence of ROS in the cells. Nitric oxide (NO) is a small lipophilic radical which diffuses across biological membranes. It is an important molecule for signalization in biological organisms. In mammals, NO notably controls blood pressure and acts as a messenger in the central nervous system [8]. Despite its role as a signaling molecule, NO is toxic for biological organisms at high concentrations, and this molecule is synthetized by macrophages to combat pathogens during the immune response through their inducible nitric oxide synthases (iNOS), making resistance to RNS critical for pathogens [9][10]. NO is more reactive than oxygen regarding the structural components of biological organisms, particularly proteins. Its toxicity comes from its ability to inhibit haem enzymes binding dioxygen, react with Fe-S centers, and indirectly induce the nitrosation of proteins [11]. Similar to oxygen, NO is relatively unreactive with most biological molecules. Direct biological targets of NO are limited to radicals and metal complexes, especially Fe-containing complexes. However, these reactions with intracellular molecules can generate other reactive species much more harmful to the cell. For example, NO with O2•− reacts at a diffusion-controlled rate to form the peroxynitrite anion (ONOO−) and its conjugated acid, peroxynitrous acid (ONOOH) (Figure 1) [12]. Since these two species coexist and are in an acid-base equilibrium in common biological conditions, the term peroxynitrite is generally used to describe both species [13]. Peroxynitrite is an impactful biological oxidant. ONOOH isomerizes to nitrate at rates of 0.095 s−1, 1.3 s−1, and 4.5 s−1 at 5 °C, 25 °C, and 37 °C, respectively [14] through the formation of both OH• and NO2•, hereafter simply noted OH and NO2 [13]. However, in biological conditions, peroxynitrite reacts with its biological targets, such as proteins, lipids, and nucleic acids, at a much faster rate, making this reaction irrelevant. Due to its high reactivity, peroxynitrite-induced damages will mostly depend on the kinetics of the reactions between peroxynitrite and its surrounding targets in priority protein metal centers, thiols, and selenols [13]. Another important RNS is NO2. This may appear alongside OH, in an aqueous environment, through the isomerization of ONOOH (Figure 1). In an air polluted by NO, such as smog, for instance, NO autoxidation in the gaseous phase leads to the formation of NO2 through the equation 2NO+O2 → 2NO2. Further reactions between NO and NO2 leads to the formation of N2O3 through the equation NO2+NO ⇋ N2O3 [15]. In aqueous media, such reactions may occur too fast to produce a significant amount of NO2 intermediate. However, these reactions still have biological relevancy as they may take place in non-aqueous media such as the lipid bilayers of membranes [16].
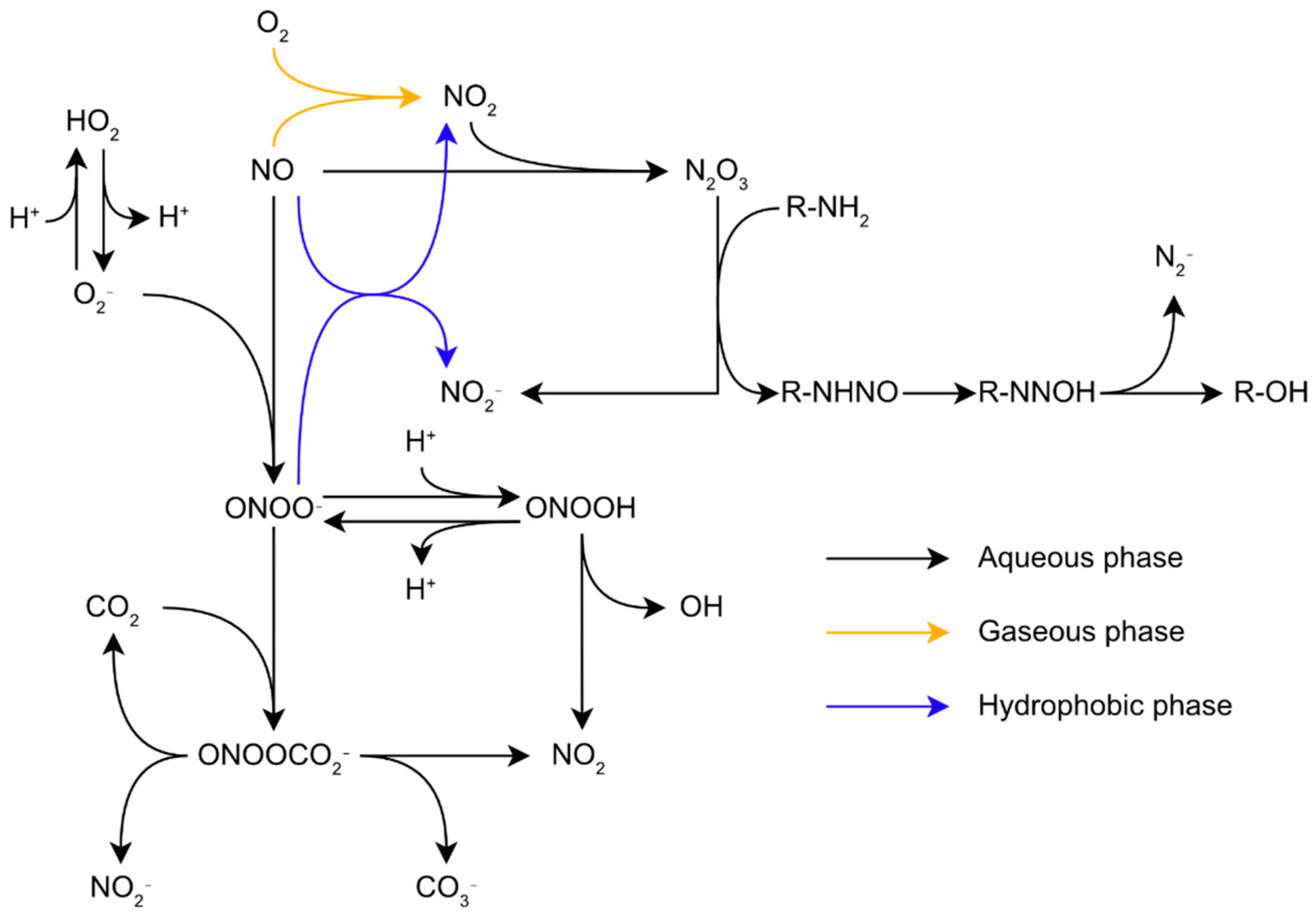
Figure 1. Reactions network of reactive oxygen and nitrogen species in biological conditions.
3. Exogenous Sources of Oxidative/Nitrosative Stress
3.1. Biotic Stress Sources
In addition to the endogenous ROS and RNS synthesis through metabolic processes described above, bacteria have to deal with exogenous stresses from various sources.
The most studied one is the oxidative burst released by phagocytic cells during the immune response. During this process, macrophages and neutrophils cause an oxidative burst on phagocyted bacteria while they synthetize O2•− through the activity of the membrane nicotinamide adenine dinucleotide (NADPH)-oxidase. Half of the O2•− reacts with H+ to form H2O2. In addition to ROS, macrophages also release large quantities of NO, synthetized by their iNOS. The simultaneous release of O2•− and NO in biological conditions lead to the formation of peroxynitrite [13]. The mammalian immune system also induces oxidative stress inside bacteria using PG Recognition Proteins (PGRPs). These proteins kill bacteria by induction of oxidative, thiol, and metal stresses in bacteria. PGRPs induce oxidative stress by blocking the bacterial respiratory chain, which promotes the production of H2O2 inside the cell [17]. In the context of host infection, bacteria are also exposed to other envelope-damaging factors such as the membrane attack complex and lysozyme, which synergize to degrade bacterial envelopes [18]. In addition to its role in bacterial killing through peptidoglycan hydrolysis, lysozyme possesses immune-dampening properties. It is notably able to decrease the neutrophil oxidative burst [19] and neutralize the prooxidant advanced glycation end products [20], which block its bactericidal activity [21].
The use of ROS as a weapon against microorganisms is not exclusive to the animal immune system. Some plants and microorganisms excrete ROS or redox-cycling compounds to suppress the growth of their competitors. For example, Gram-positive lactic acid bacteria are able to release important concentrations of H2O2 in their environment. In fact, they lack full respiratory chains, but many of them still use oxygen as a direct electron acceptor, thanks to lactate and pyruvate oxidases [22][23]. The obtained product is converted by an acetate kinase in another ATP molecule(s), as well as H2O2. In favorable experimental conditions, lactic acid bacteria can produce millimolar concentrations of H2O2, inhibiting the growth of other bacteria [22][24][25]. However, in natural conditions, H2O2 might be carried away or otherwise degraded; therefore, such concentrations might not be reached. Another example is the production of pyocyanin by P. aeruginosa. This secondary metabolite is able to inhibit the respiration chain, leading to the impairment of energy-dependent transport systems and the production of oxidative species, such as H2O2 [26][27].
Another source of oxidative stress could be antibiotics. Oxidative stress is not directly the primary mode of action of antibiotics to suppress bacterial growth, although new antimicrobial relying on the production of ROS and RNS are gaining interest [28]. Indeed, antibiotics mainly target peptidoglycan biosynthesis, protein synthesis, or DNA replication and repair. However, oxidative stress could be a secondary effect of some antibiotics. For instance, some bactericidal antibiotics [29][30] could lead to the intracellular production of ROS such as O2•− and H2O2 [28][31][32], although these results have been challenged [29][30]. Furthermore, the response to some antibiotics depends on their species [33]. Overall, the impact of oxidative stress in the modes of action of traditional antibiotics on Gram-negative bacteria is a complex question requiring further clarification. In addition to their oxidative potential, antibiotics often induce protective responses leading to the reshaping of the envelope, especially by modulating the efflux pumps and porins at the outer membrane through the triggering of envelope regulators, such as the Cpx complex [34].
3.2. Abiotic Stress Sources
Environmental bacteria are also subjected to oxidative and nitrosative stresses through multiple abiotic sources. An important source is ultraviolet (UV) radiations, which can induce oxidative stress inside the cells by direct exposure or through the generation of H2O2 in surface water through UV photochemistry [35][36][37]. Anthropogenic activities also create the conditions for oxidative and nitrosative stresses in the environment. The most glaring example is the release of vast amounts of ROS and RNS into the atmosphere by fuel combustion processes used in transport and industry [38][39], which can affect bacterial physiology [40][41].
3.3. Targets of ROS and RNS
Overall, the exposure of bacteria to excessive oxidative and nitrosative stresses destabilize the envelope, impairing its proper functions, inducing membrane permeabilization [41] and hyperpolarization [42] and eventually leading to cell death. The harm caused by ROS and RNS to the bacterial envelope comes from their ability to react with biomolecules and alter their biochemical properties, disturbing the biochemical processes necessary for cell survival. The vast range of ROS and RNS leads to a wide panel of biological targets. In the envelope, the main targets for oxidative and nitrosative stresses are proteins, and in some cases, phospholipids. These molecules can undergo various alterations discussed below, constituting a real challenge for cells to maintain their envelope integrity.
3.3.1. Phospholipids
ROS and RNS are relatively unreactive with the phospholipids forming biological membranes, except for phospholipids containing polyunsaturated fatty acids (PUFAs), on which ROS and RNS induce their lipid peroxidation. Most bacteria do not synthetize PUFAs, and as a result, most bacterial membranes are composed of saturated or monounsaturated phospholipids [43], thence theoretically insensitive to ROS and RNS. However, studies on P. fluorescens showed modifications of the membrane phospholipid composition after exposure to NO2, despite the absence of PUFAs in the strain [40][41]. By contrast, in various water sources and fish microbiota, bacteria, mostly from the Shewanella genus, synthetize PUFAs, such as eicosapentaenoic acid and docosahexaenoic acid, which are prone to peroxidation [44]. Moreover, even bacteria that are unable to synthetize them may incorporate PUFAs from their environment into their membranes. For example, various Vibrio species, such as V. cholerae, V. vulnificus, and V. parahaemolyticus, possess the mechanisms required to accumulate and incorporate PUFAs into their membranes [45].
Noticeably, the vertebrate immune system takes advantage of such a PUFA incorporation mechanism to initiate lipid peroxidation in bacteria. In vertebrates, arachidonic acid is released concomitantly with RNS and ROS during the oxidative burst. In the context of the immune response, PUFAs are toxic for a wide range of bacteria such as Acinetobacter baumannii [46], Listeria monocytogenes [47], Pseudomonas aeruginosa [47], including gram-positive species, such as Staphylococcus aureus [48], Cutibacterium acnes [48], and Streptococcus pneumoniae [49]. The PUFAs toxicity depends on their capacity to esterify the fatty acids into the bacterial membranes [50]. In the case of the Gram-positive Staphylococcus aureus, the toxicity of PUFAs was shown to be specifically mediated by lipid peroxidation [51]. As a result, even though bacteria are relatively less sensitive to lipid peroxidation than other genera, this process still plays a crucial role in bacterial pathogenicity.
3.3.2. Peptidoglycan
Although the effects of oxidative and nitrosative stress on peptidoglycan are poorly understood, a few ones were reported links between oxidative or nitrosative stress and proteins associated with peptidoglycan or its synthesis [52][53][54][55]. The clearest demonstrated effect of oxidative stress on peptidoglycan comes from a recent one by Giacomucci et al. It was showed that the absence of the protein ElyC of unknown function induces the overproduction of OH• in the periplasm, which leads to a direct or indirect interruption of peptidoglycan synthesis [56].
3.3.3. Envelope Proteins
ROS and RNS can react with a wide variety of protein features and virtually with any amino acid. However, the susceptibility of amino acids to ROS and RNS oxidation varies, and the reactions taking place are determined by their reaction rate. ROS react especially well with the amino acids containing sulfur, methionine, and cysteine [57]. The oxidation of methionine residues by ROS forms one of two diastereoisomers of methionine sulfoxide (Met-O): methionine-S-sulfoxide or methionine-R-sulfoxide. None of the couple of Met-O stereoisomers is preferably formed over the other, but different reductases are required to reduce each one [58]. ROS oxidation of cysteine residues first generates a highly reactive sulfenic acid derivative (RSOH). Depending on its microenvironment, RSOH may also further react with a nearby cysteine to form a disulfide bond or be oxidized to sulfinic (RSO2H) and sulfonic (RSO3H) acid. Sulfenic acid and methionine sulfoxide formation may induce misfolding, inactivation, or degradation of proteins [59][60].
3.3.4. Protein Carbonylation
ROS can also induce irreversible protein modifications through protein carbonylation. So ROS react with the side chains of amino acids to form carbonyl groups [61]. Protein carbonylation is directly induced by ROS such as H2O2 on the side chains of amino acids: arginine, lysine, proline, and threonine [62]. Furthermore, aldehydes formed by lipid peroxidation can also indirectly target the side chains of lysine, histidine, and cysteine through the Michael reaction [63][64]. Similar to methionine sulfoxide and sulfenic acid, carbonylation can impair protein functions. Protein carbonyls are more difficult to induce than methionine sulfoxide and sulfenic acid and then are considered markers of a more intense oxidative stress [65]. In E. coli, for example, the entry into the stationary phase caused by nitrogen or carbon starvation leads to an increase in protein carbonylation [66].
3.3.5. Protein S-Nitrosylation
Cysteines are also prone to S-nitrosylation, a process by which NO moieties covalently bind to thiols to form an S-nitrosothiol (SNO). Its formation is related to reaction with different RNS, such as NO and N2O3, or NO carriers such as other nitrosothiols. This process may alter protein function. Protein S-nitrosylation is reversible and is often enzymatically mediated by organisms to protect their proteins against untargeted S-nitrosylation from exogenous nitrosative stress [67].
3.3.6. Tyrosine Nitration
The most characteristic protein modification induced by peroxynitrite is tyrosine nitration. Its first step consists of the one-electron oxidation of the tyrosine phenolic ring to form a tyrosine radical, Tyr•. This step is performed by various ROS and RNS, such as OH, NO2, or secondary produces of peroxynitrite reactions, CO3−, and oxo-metal complexes. In hydrophobic environments, tyrosine nitration may also be initiated by the intermediates of lipid peroxidation, such as lipid peroxyl LOO• and alkoxyl LO• [68][69]. These radicals contribute to tyrosine nitration within lipid bilayers. During the second step of the tyrosine nitration, Tyr• reacts with NO2 to form NO2Tyr. The complete process impairs protein function and leads to loss of function but is compensated by a gain in proteins implicated in regulatory cascades [13].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10050924
References
- Ligeza, A.; Tikhonov, A.N.; Hyde, J.S.; Subczynski, W.K. Oxygen permeability of thylakoid membranes: Electron paramagnetic resonance spin labeling study. Biochim. Biophys. Acta 1998, 1365, 453–463.
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454.
- Korshunov, S.; Imlay, J.A. Two sources of endogenous H2O2 in Escherichia coli. Mol. Microbiol. 2010, 75, 1389–1401.
- Bielski, B.H.J. Reevaluation of the Spectral and Kinetic Properties of Ho2 and O2- Free Radicals. Photochem. Photobiol. 1978, 28, 645–649.
- McCord, J.M.; Fridovich, I. Superoxide Dismutase: An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055.
- Panov, A.V.; Dikalov, S.I. Cardiolipin, Perhydroxyl Radicals, and Lipid Peroxidation in Mitochondrial Dysfunctions and Aging. Oxid. Med. Cell. Longev. 2020, 2020, 1323028.
- Glaeser, J.; Nuss, A.M.; Berghoff, B.A.; Klug, G. Chapter 4—Singlet Oxygen Stress in Microorganisms. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 58, pp. 141–173.
- Garcia, X.; Stein, F. Nitric Oxide. Semin. Pediatr. Infect. Dis. 2006, 17, 55–57.
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vazquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143.
- Flint, A.; Stintzi, A.; Saraiva, L.M. Oxidative and nitrosative stress defences of Helicobacter and Campylobacter species that counteract mammalian immunity. FEMS Microbiol. Rev. 2016, 40, 938–960.
- Poole, R.K. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 2005, 33, 176–180.
- Hughes, M.N. Relationships between nitric oxide, nitroxyl ion, nitrosonium cation and peroxynitrite. Biochim. Biophys. Acta BBA Bioenerg. 1999, 1411, 263–272.
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of Peroxynitrite and Protein Tyrosine Nitration. Chem. Rev. 2018, 118, 1338–1408.
- Koppenol, W.H.; Moreno, J.J.; Pryor, W.A.; Ischiropoulos, H.; Beckman, J.S. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 1992, 5, 834–842.
- Wink, D.A.; Ford, P.C. Nitric Oxide Reactions Important to Biological Systems: A Survey of Some Kinetics Investigations. Methods 1995, 7, 14–20.
- Hughes, M.N. Chapter One—Chemistry of Nitric Oxide and Related Species. In Methods in Enzymology; Poole, R.K., Ed.; Globins and Other Nitric Oxide-Reactive Proteins, Part A; Academic Press: Cambridge, MA, USA, 2008; Volume 436, pp. 3–19.
- Kashyap, D.R.; Kuzma, M.; Kowalczyk, D.A.; Gupta, D.; Dziarski, R. Bactericidal peptidoglycan recognition protein induces oxidative stress in Escherichia coli through a block in respiratory chain and increase in central carbon catabolism. Mol. Microbiol. 2017, 105, 755–776.
- Heesterbeek, D.A.C.; Muts, R.M.; van Hensbergen, V.P.; de Saint Aulaire, P.; Wennekes, T.; Bardoel, B.W.; van Sorge, N.M.; Rooijakkers, S.H.M. Outer membrane permeabilization by the membrane attack complex sensitizes Gram-negative bacteria to antimicrobial proteins in serum and phagocytes. PLOS Pathog. 2021, 17, e1009227.
- Liu, H.; Zheng, F.; Cao, Q.; Ren, B.; Zhu, L.; Striker, G.; Vlassara, H. Amelioration of oxidant stress by the defensin lysozyme. Am. J. Physiol.-Endocrinol. Metab. 2006, 290, E824–E832.
- Li, Y.M.; Tan, A.X.; Vlassara, H. Antibacterial activity of lysozyme and lactoferrin is inhibited by binding of advanced glycation–modified proteins to a conserved motif. Nat. Med. 1995, 1, 1057–1061.
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512.
- Seki, M.; Iida, K.; Saito, M.; Nakayama, H.; Yoshida, S. Hydrogen peroxide production in Streptococcus pyogenes: Involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 2004, 186, 2046–2051.
- Spellerberg, B.; Cundell, D.R.; Sandros, J.; Pearce, B.J.; Idanpaan-Heikkila, I.; Rosenow, C.; Masure, H.R. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 1996, 19, 803–813.
- Liu, X.; Ramsey, M.M.; Chen, X.; Koley, D.; Whiteley, M.; Bard, A.J. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 2668–2673.
- Tong, H.; Chen, W.; Merritt, J.; Qi, F.; Shi, W.; Dong, X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: A possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 2007, 63, 872–880.
- Baron, S.S.; Terranova, G.; Rowe, J.J. Molecular mechanism of the antimicrobial action of pyocyanin. Curr. Microbiol. 1989, 18, 223–230.
- Hassan, H.M.; Fridovich, I. Mechanism of the antibiotic action pyocyanine. J. Bacteriol. 1980, 141, 156–163.
- Mourenza, Á.; Gil, J.A.; Mateos, L.M.; Letek, M. Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance. Antioxidants 2020, 9, 361.
- Keren, I.; Wu, Y.; Inocencio, J.; Mulcahy, L.R.; Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013, 339, 1213–1216.
- Mahoney, T.F.; Silhavy, T.J. The Cpx Stress Response Confers Resistance to Some, but Not All, Bactericidal Antibiotics. J. Bacteriol. 2013, 195, 1869–1874.
- Albesa, I.; Becerra, M.C.; Battán, P.C.; Páez, P.L. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 2004, 317, 605–609.
- Goswami, M.; Mangoli, S.H.; Jawali, N. Involvement of Reactive Oxygen Species in the Action of Ciprofloxacin against Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 949–954.
- Ladjouzi, R.; Bizzini, A.; Lebreton, F.; Sauvageot, N.; Rincé, A.; Benachour, A.; Hartke, A. Analysis of the tolerance of pathogenic Enterococci and Staphylococcus aureus to cell wall active antibiotics. J. Antimicrob. Chemother. 2013, 68, 2083–2091.
- Dam, S.; Pagès, J.-M.; Masi, M. Stress responses, outer membrane permeability control and antimicrobial resistance in Enterobacteriaceae. Microbiol. Read. Engl. 2018, 164, 260–267.
- Lesser, M.P. Oxidative stress in marine environments: Biochemistry and Physiological Ecology. Annu. Rev. Physiol. 2006, 68, 253–278.
- Meslé, M.M.; Beam, J.P.; Jay, Z.J.; Bodle, B.; Bogenschutz, E.; Inskeep, W.P. Hydrogen Peroxide Cycling in High-Temperature Acidic Geothermal Springs and Potential Implications for Oxidative Stress Response. Front. Mar. Sci. 2017, 4, 130.
- Santos, A.L.; Gomes, N.C.M.; Henriques, I.; Almeida, A.; Correia, A.; Cunha, Â. Growth conditions influence UVB sensitivity and oxidative damage in an estuarine bacterial isolate. Photochem. Photobiol. Sci. 2013, 12, 974–986.
- Depayras, S.; Kondakova, T.; Heipieper, H.J.; Feuilloley, M.G.; Orange, N.; Duclairoir-Poc, C. The Hidden Face of Nitrogen Oxides Species: From Toxic Effects to Potential Cure? IntechOpen: London, UK, 2018; ISBN 978-1-78923-385-8.
- Skalska, K.; Miller, J.S.; Ledakowicz, S. Trends in NOx abatement: A review. Sci. Total Environ. 2010, 408, 3976–3989.
- Chautrand, T.; Souak, D.; Kondakova, T.; Depayras, S.; Machour, N.; Heipieper, H.; Feuilloley, M.; Orange, N.; Poc, C. Air pollution and other environmental stresses: Gaseous NO2 exposure leads to specific alterations of Pseudomonas fluorescens. WIT Trans. Ecol. Environ. 2020, 244, 53–63.
- Depayras, S.; Kondakova, T.; Merlet-Machour, N.; Heipieper, H.J.; Barreau, M.; Catovic, C.; Feuilloley, M.; Orange, N.; Poc, C.D. Impact of gaseous NO2 on P. fluorescens strain in the membrane adaptation and virulence. Int. J. Environ. Impacts 2018, 1, 183–192.
- Blee, J.A.; Roberts, I.S.; Waigh, T.A. Membrane potentials, oxidative stress and the dispersal response of bacterial biofilms to 405 nm light. Phys. Biol. 2020, 17, 036001.
- Strahl, H.; Errington, J. Bacterial Membranes: Structure, Domains, and Function. Annu. Rev. Microbiol. 2017, 71, 519–538.
- Dailey, F.E.; McGraw, J.E.; Jensen, B.J.; Bishop, S.S.; Lokken, J.P.; Dorff, K.J.; Ripley, M.P.; Munro, J.B. The Microbiota of Freshwater Fish and Freshwater Niches Contain Omega-3 Fatty Acid-Producing Shewanella Species. Appl. Environ. Microbiol. 2015, 82, 218–231.
- Moravec, A.R.; Siv, A.W.; Hobby, C.R.; Lindsay, E.N.; Norbash, L.V.; Shults, D.J.; Symes, S.J.K.; Giles, D.K. Exogenous Polyunsaturated Fatty Acids Impact Membrane Remodeling and Affect Virulence Phenotypes among Pathogenic Vibrio Species. Appl. Environ. Microbiol. 2017, 83, e01415-17.
- Jiang, J.-H.; Hassan, K.A.; Begg, S.L.; Rupasinghe, T.W.T.; Naidu, V.; Pederick, V.G.; Khorvash, M.; Whittall, J.J.; Paton, J.C.; Paulsen, I.T.; et al. Identification of Novel Acinetobacter baumannii Host Fatty Acid Stress Adaptation Strategies. mBio 2019, 10, e02056-18.
- Shin, S.Y.; Bajpai, V.K.; Kim, H.R.; Kang, S.C. Antibacterial activity of bioconverted eicosapentaenoic (EPA) and docosahexaenoic acid (DHA) against foodborne pathogenic bacteria. Int. J. Food Microbiol. 2007, 113, 233–236.
- Desbois, A.P.; Lawlor, K.C. Antibacterial Activity of Long-Chain Polyunsaturated Fatty Acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs 2013, 11, 4544–4557.
- Eijkelkamp, B.A.; Begg, S.L.; Pederick, V.G.; Trapetti, C.; Gregory, M.K.; Whittall, J.J.; Paton, J.C.; McDevitt, C.A. Arachidonic Acid Stress Impacts Pneumococcal Fatty Acid Homeostasis. Front. Microbiol. 2018, 9, 813.
- Krute, C.N.; Ridder, M.J.; Seawell, N.A.; Bose, J.L.Y. Inactivation of the exogenous fatty acid utilization pathway leads to increased resistance to unsaturated fatty acids in Staphylococcus aureus. Microbiology 2021, 165, 197–207.
- Beavers, W.N.; Monteith, A.J.; Amarnath, V.; Mernaugh, R.L.; Roberts, L.J.; Chazin, W.J.; Davies, S.S.; Skaar, E.P. Arachidonic Acid Kills Staphylococcus aureus through a Lipid Peroxidation Mechanism. mBio 2019, 10, e01333-19.
- Thibessard, A.; Fernandez, A.; Gintz, B.; Leblond-Bourget, N.; Decaris, B. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophilus CNRZ368. J. Bacteriol. 2002, 184, 2821–2826.
- Wang, G.; Olczak, A.; Forsberg, L.S.; Maier, R.J. Oxidative Stress-induced Peptidoglycan Deacetylase in Helicobacter pylori. J. Biol. Chem. 2009, 284, 6790–6800.
- Davis, M.M.; Brock, A.M.; DeHart, T.G.; Boribong, B.P.; Lee, K.; McClune, M.E.; Chang, Y.; Cramer, N.; Liu, J.; Jones, C.N.; et al. The peptidoglycan-associated protein NapA plays an important role in the envelope integrity and in the pathogenesis of the lyme disease spirochete. PLoS Pathog. 2021, 17, e1009546.
- Chautrand, T.; Depayras, S.; Souak, D.; Kondakova, T.; Barreau, M.; Kentache, T.; Hardouin, J.; Tahrioui, A.; Thoumire, O.; Konto-Ghiorghi, Y.; et al. Gaseous NO2 induces various envelope alterations in Pseudomonas fluorescens MFAF76a. Sci. Rep. 2022; in press.
- Giacomucci, S.; Alvarez, L.; Rodrigues, C.D.A.; Cava, F.; Paradis-Bleau, C. Hydroxyl Radical Overproduction in the Envelope: An Achilles’ Heel in Peptidoglycan Synthesis. Microbiol. Spectr. 2022, 10, e01203-21.
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396.
- Lee, B.C.; Gladyshev, V.N. The biological significance of methionine sulfoxide stereochemistry. Free Radic. Biol. Med. 2011, 50, 221–227.
- Davies, M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta 2005, 1703, 93–109.
- Roos, G.; Messens, J. Protein sulfenic acid formation: From cellular damage to redox regulation. Free Radic. Biol. Med. 2011, 51, 314–326.
- Suzuki, Y.J.; Carini, M.; Butterfield, D.A. Protein Carbonylation. Antioxid. Redox Signal. 2010, 12, 323–325.
- Stadtman, E.R.; Levine, R.L. Protein oxidation. Ann. N. Y. Acad. Sci. 2000, 899, 191–208.
- Grimsrud, P.A.; Xie, H.; Griffin, T.J.; Bernlohr, D.A. Oxidative Stress and Covalent Modification of Protein with Bioactive Aldehydes. J. Biol. Chem. 2008, 283, 21837–21841.
- Sayre, L.M.; Lin, D.; Yuan, Q.; Zhu, X.; Tang, X. Protein adducts generated from products of lipid oxidation: Focus on HNE and one. Drug Metab. Rev. 2006, 38, 651–675.
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation in human diseases. Trends Mol. Med. 2003, 9, 169–176.
- Ballesteros, M.; Fredriksson, Å.; Henriksson, J.; Nyström, T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001, 20, 5280–5289.
- Seth, D.; Hausladen, A.; Stamler, J.S. Anaerobic Transcription by OxyR: A Novel Paradigm for Nitrosative Stress. Antioxid. Redox Signal. 2020, 32, 803–816.
- Bartesaghi, S.; Wenzel, J.; Trujillo, M.; López, M.; Joseph, J.; Kalyanaraman, B.; Radi, R. Lipid peroxyl radicals mediate tyrosine dimerization and nitration in membranes. Chem. Res. Toxicol. 2010, 23, 821–835.
- Folkes, L.K.; Bartesaghi, S.; Trujillo, M.; Radi, R.; Wardman, P. Kinetics of oxidation of tyrosine by a model alkoxyl radical. Free Radic. Res. 2012, 46, 1150–1156.
This entry is offline, you can click here to edit this entry!
