Living beings spend their lives and carry out their daily activities interacting with environmental situations that present space-time variations and that involve contact with other life forms, which may behave as commensals or as invaders and/or parasites. The characteristics of the environment, as well as the processes that support the maintenance of life and that characterize the execution of activities of daily life generally present periodic variations, which are mostly synchronized with the light–dark cycle determined by Earth’s rotation on its axis. These rhythms with 24-h periodicity, defined as circadian, influence events linked to the interaction between hosts and hosted microorganisms and can dramatically determine the outcome of this interplay.
- clock
- circadian rhythms
- virus
- host
- immune system
1. The Circadian Clock Circuitry
2. The Molecular Mechanisms of Biological Ticking
3. Viruses and Circadian Clock Circuits
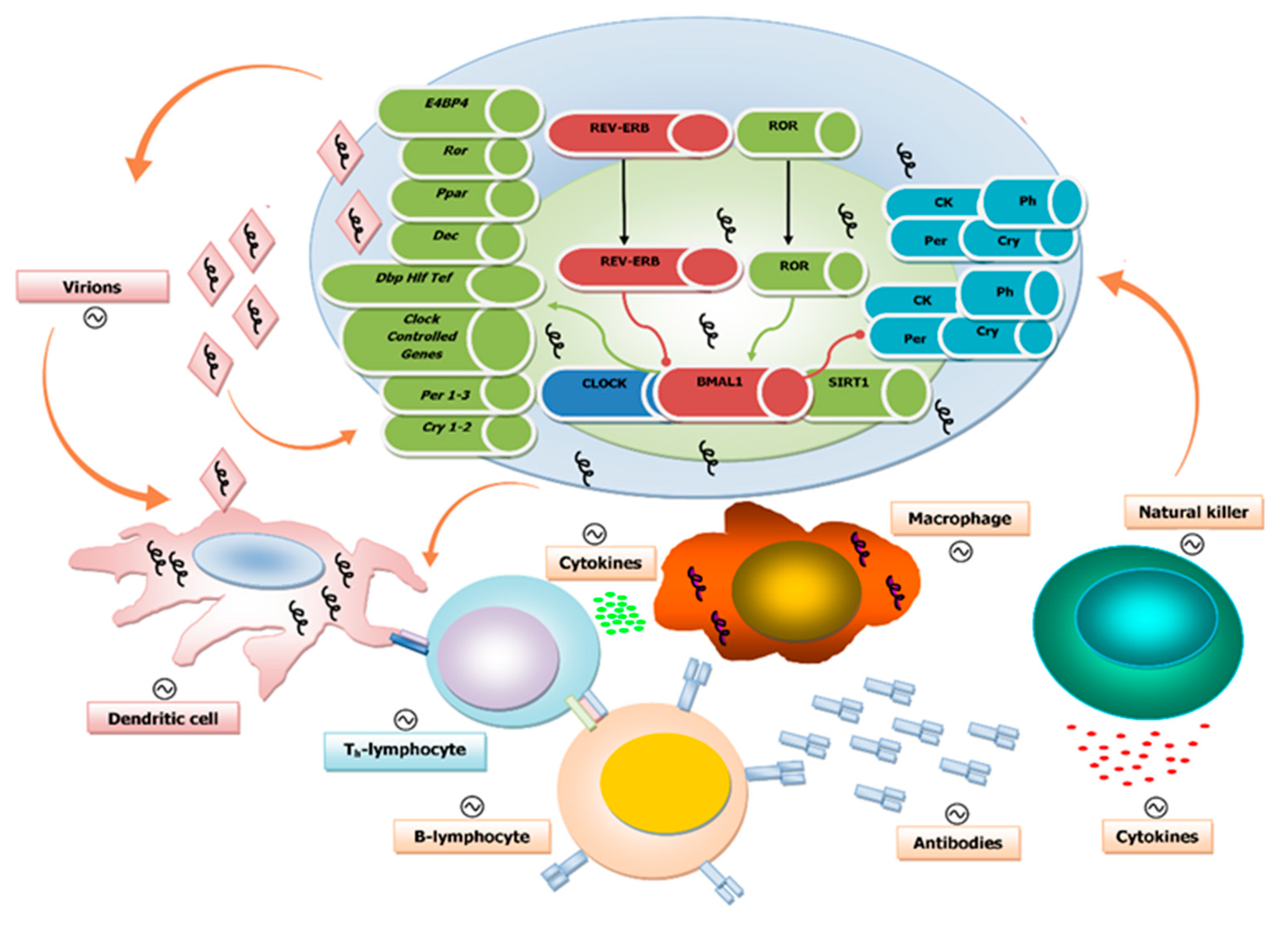
4. Circadian Regulation of Both Innate and Adaptive Immune Systems
5. Influence of the Biological Clock on Virus Replication Cycle and Disease
6. Disruption of the Circadian Clock by Viruses
7. The Biological Clock and Influenza Virus Infection
8. Epigenetic Mechanisms Underlying the Interaction Between Viral Infection and the Circadian Clock Machinery
This entry is adapted from the peer-reviewed paper 10.3390/pathogens9020083
References
- Abrahamson, E.E.; Moore, R.Y. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001, 916, 172–191.
- Stephan, F.K.; Zucker, I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. USA 1972, 69, 1583–1586.
- Moore, R.Y.; Eichler, V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972, 42, 201–206.
- Lehman, M.N.; Silver, R.; Gladstone, W.R.; Kahn, R.M.; Gibson, M.; Bittman, E.L. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci. 1987, 7, 1626–1638.
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978.
- Sujino, M.; Masumoto, K.H.; Yamaguchi, S.; van der Horst, G.T.; Okamura, H.; Inouye, S.T. Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr. Biol. 2003, 13, 664–668.
- Bjarnason, G.A.; Jordan, R.C.; Wood, P.A.; Li, Q.; Lincoln, D.W.; Sothern, R.B.; Hrushesky, W.J.; Ben-David, Y. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am. J. Pathol. 2001, 158, 1793–1801.
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685.
- Yoo, S.H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.K.; Oh, W.J.; Yoo, O.J.; et al. Period2: Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346.
- McNamara, P.; Seo, S.B.; Rudic, R.D.; Sehgal, A.; Chakravarti, D.; FitzGerald, G.A. Regulation of clock and mop4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell 2001, 105, 877–889.
- Lowrey, P.L.; Takahashi, J.S. Genetics of circadian rhythms in mammalian model organisms. Adv. Genet. 2011, 74, 175–230.
- King, D.P.; Takahashi, J.S. Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 2000, 23, 713–742.
- Antle, M.C.; Silver, R. Orchestrating time: Arrangements of the brain circadian clock. Trends Neurosci. 2005, 28, 145–151.
- Gooley, J.J.; Lu, J.; Chou, T.C.; Scammell, T.E.; Saper, C.B. Melanopsin in cells of origin of the retinohypothalamic tract. Nat. Neurosci. 2001, 4, 1165.
- Hannibal, J.; Hindersson, P.; Knudsen, S.M.; Georg, B.; Fahrenkrug, J. The photopigment melanopsin is exclusively present in pituitary adenylate cyclase-activating polypeptide-containing retinal ganglion cells of the retinohypothalamic tract. J. Neurosci. 2002, 22, RC191.
- Hannibal, J.; Hindersson, P.; Ostergaard, J.; Georg, B.; Heegaard, S.; Larsen, P.J.; Fahrenkrug, J. Melanopsin is expressed in pacap-containing retinal ganglion cells of the human retinohypothalamic tract. Invest. Ophthalmol. Vis. Sci. 2004, 45, 4202–4209.
- Hattar, S.; Liao, H.W.; Takao, M.; Berson, D.M.; Yau, K.W. Melanopsin-containing retinal ganglion cells: Architecture, projections, and intrinsic photosensitivity. Science 2002, 295, 1065–1070.
- Hannibal, J.; Moller, M.; Ottersen, O.P.; Fahrenkrug, J. Pacap and glutamate are co-stored in the retinohypothalamic tract. J. Comp. Neurol. 2000, 418, 147–155.
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941.
- Hastings, M.H.; Herzog, E.D. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J. Biol. Rhythms 2004, 19, 400–413.
- Moore, R.Y.; Speh, J.C.; Leak, R.K. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002, 309, 89–98.
- Moore, R.Y.; Speh, J.C. Gaba is the principal neurotransmitter of the circadian system. Neurosci. Lett. 1993, 150, 112–116.
- Hirota, T.; Fukada, Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog. Sci. 2004, 21, 359–368.
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179.
- Reddy, A.B.; O’Neill, J.S. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010, 20, 36–44.
- Staels, B. When the clock stops ticking, metabolic syndrome explodes. Nat. Med. 2006, 12, 54–55.
- Baraldo, M. The influence of circadian rhythms on the kinetics of drugs in humans. Expert Opin. Drug Metab. Toxicol. 2008, 4, 175–192.
- Levi, F. Therapeutic implications of circadian rhythms in cancer patients. Novartis Found. Symp. 2000, 227, 119–136.
- Levi, F.; Schibler, U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 593–628.
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198.
- Fortier, E.E.; Rooney, J.; Dardente, H.; Hardy, M.P.; Labrecque, N.; Cermakian, N. Circadian variation of the response of t cells to antigen. J. Immunol. 2011, 187, 6291–6300.
- Gibbs, J.; Ince, L.; Matthews, L.; Mei, J.; Bell, T.; Yang, N.; Saer, B.; Begley, N.; Poolman, T.; Pariollaud, M.; et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 2014, 20, 919–926.
- Nguyen, K.D.; Fentress, S.J.; Qiu, Y.; Yun, K.; Cox, J.S.; Chawla, A. Circadian gene bmal1 regulates diurnal oscillations of ly6c(hi) inflammatory monocytes. Science 2013, 341, 1483–1488.
- Silver, A.C.; Arjona, A.; Walker, W.E.; Fikrig, E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 2012, 36, 251–261.
- Pick, R.; He, W.; Chen, C.S.; Scheiermann, C. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol. 2019, 40, 524–537.
- Arjona, A.; Sarkar, D.K. Evidence supporting a circadian control of natural killer cell function. Brain Behav. Immun. 2006, 20, 469–476.
- Bollinger, T.; Leutz, A.; Leliavski, A.; Skrum, L.; Kovac, J.; Bonacina, L.; Benedict, C.; Lange, T.; Westermann, J.; Oster, H.; et al. Circadian clocks in mouse and human CD4+ T cells. PLoS ONE 2011, 6, e29801.
- Keller, M.; Mazuch, J.; Abraham, U.; Eom, G.D.; Herzog, E.D.; Volk, H.D.; Kramer, A.; Maier, B. A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. USA 2009, 106, 21407–21412.
- Froy, O.; Chapnik, N. Circadian oscillation of innate immunity components in mouse small intestine. Mol. Immunol. 2007, 44, 1954–1960.
- Druzd, D.; Matveeva, O.; Ince, L.; Harrison, U.; He, W.; Schmal, C.; Herzel, H.; Tsang, A.H.; Kawakami, N.; Leliavski, A.; et al. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 2017, 46, 120–132.
- Ehlers, A.; Xie, W.; Agapov, E.; Brown, S.; Steinberg, D.; Tidwell, R.; Sajol, G.; Schutz, R.; Weaver, R.; Yu, H.; et al. Bmal1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal. Immunol. 2018, 11, 97–111.
- Majumdar, T.; Dhar, J.; Patel, S.; Kondratov, R.; Barik, S. Circadian transcription factor bmal1 regulates innate immunity against select RNA viruses. Innate Immun. 2017, 23, 147–154.
- Phillips, A.C.; Gallagher, S.; Carroll, D.; Drayson, M. Preliminary evidence that morning vaccination is associated with an enhanced antibody response in men. Psychophysiology 2008, 45, 663–666.
- Long, J.E.; Drayson, M.T.; Taylor, A.E.; Toellner, K.M.; Lord, J.M.; Phillips, A.C. Morning vaccination enhances antibody response over afternoon vaccination: A cluster-randomised trial. Vaccine 2016, 34, 2679–2685.
- Kirby, T. Influenza vaccination in the morning improves response. Lancet. Respir. Med. 2016, 4, 435.
- Dowell, S.F. Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg. Infect. Dis. 2001, 7, 369–374.
- Dopico, X.C.; Evangelou, M.; Ferreira, R.C.; Guo, H.; Pekalski, M.L.; Smyth, D.J.; Cooper, N.; Burren, O.S.; Fulford, A.J.; Hennig, B.J.; et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 2015, 6, 7000.
- Edgar, R.S.; Stangherlin, A.; Nagy, A.D.; Nicoll, M.P.; Efstathiou, S.; O’Neill, J.S.; Reddy, A.B. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. USA 2016, 113, 10085–10090.
- Matsuzawa, T.; Nakamura, Y.; Ogawa, Y.; Ishimaru, K.; Goshima, F.; Shimada, S.; Nakao, A.; Kawamura, T. Differential day-night outcome to HSV-2 cutaneous infection. J. Invest. Dermatol. 2018, 138, 233–236.
- Huitron-Resendiz, S.; Marcondes, M.C.; Flynn, C.T.; Lanigan, C.M.; Fox, H.S. Effects of simian immunodeficiency virus on the circadian rhythms of body temperature and gross locomotor activity. Proc. Natl. Acad. Sci. USA 2007, 104, 15138–15143.
- Yang, S.L.; Yu, C.; Jiang, J.X.; Liu, L.P.; Fang, X.; Wu, C. Hepatitis B virus X protein disrupts the balance of the expression of circadian rhythm genes in hepatocellular carcinoma. Oncol. Lett. 2014, 8, 2715–2720.
- Sahar, S.; Sassone-Corsi, P. The epigenetic language of circadian clocks. Handb. Exp. Pharmacol. 2013, 29–44.
- Jueliger, S.; Lyons, J.; Cannito, S.; Pata, I.; Pata, P.; Shkolnaya, M.; Lo Re, O.; Peyrou, M.; Villarroya, F.; Pazienza, V.; et al. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics 2016, 11, 709–720.
- Benegiamo, G.; Vinciguerra, M.; Mazzoccoli, G.; Piepoli, A.; Andriulli, A.; Pazienza, V. DNA methyltransferases 1 and 3b expression in huh-7 cells expressing HCV core protein of different genotypes. Dig. Dis. Sci. 2012, 57, 1598–1603.
- Ripoli, M.; Barbano, R.; Balsamo, T.; Piccoli, C.; Brunetti, V.; Coco, M.; Mazzoccoli, G.; Vinciguerra, M.; Pazienza, V. Hypermethylated levels of e-cadherin promoter in huh-7 cells expressing the HCV core protein. Virus Res. 2011, 160, 74–81.
