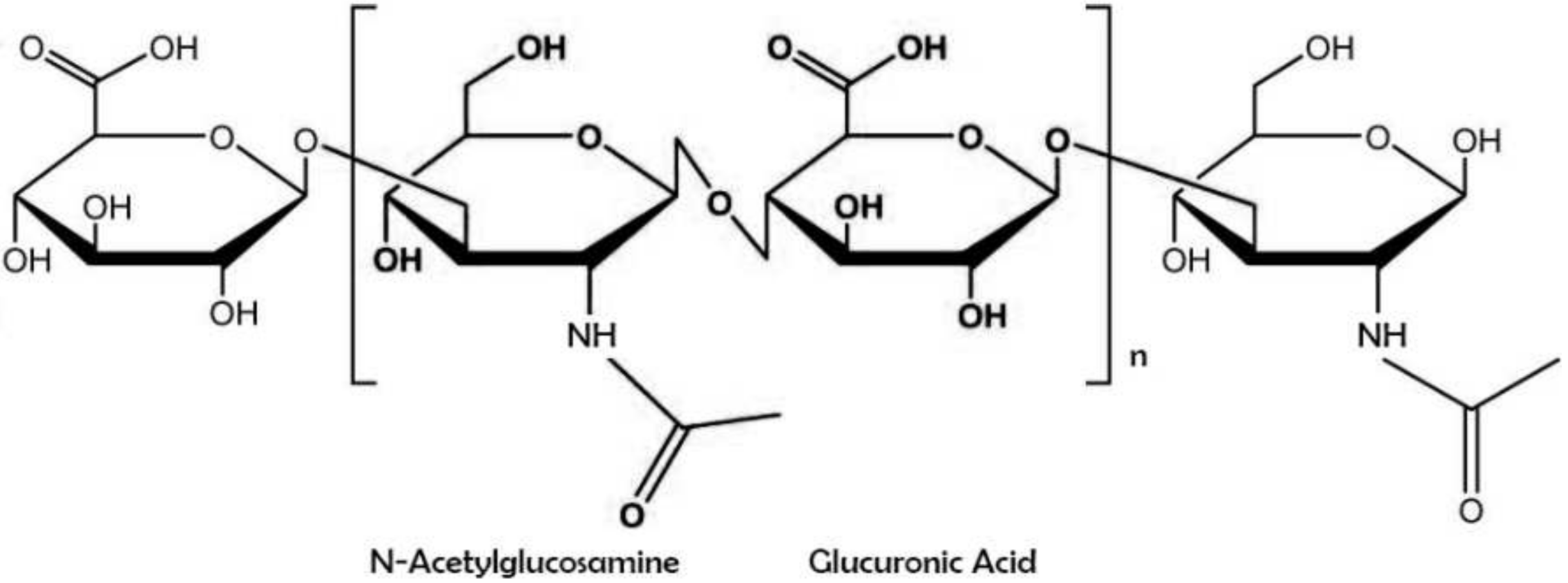
Hyaluronic acid (also known as sodium hyaluronate or hyaluronan) is a straight-chain, natural polysaccharide and the only nonsulfated GAG composed of alternating (1–4)-β d-glucuronic and (1–3)-β N-acetyl-d-glucosamine units. Both carbohydrate units are spatially related to glucose; therefore, in the β-configuration, it is possible for all their bulky groups (hydroxyl and carboxyl groups and the anomeric carbon on the neighboring sugar) to be in sterically favorable planes, while all the small hydrogen atoms occupy less sterically favorable axial positions. This chemical structure of HA is energetically very stable because of interactions between hydrophobic and intermolecular hydrogen bonds and the acetamide and carboxylate groups.
- hyaluronic acid
- synthesis
- degradation
1. Introducing Hyaluronic Acid
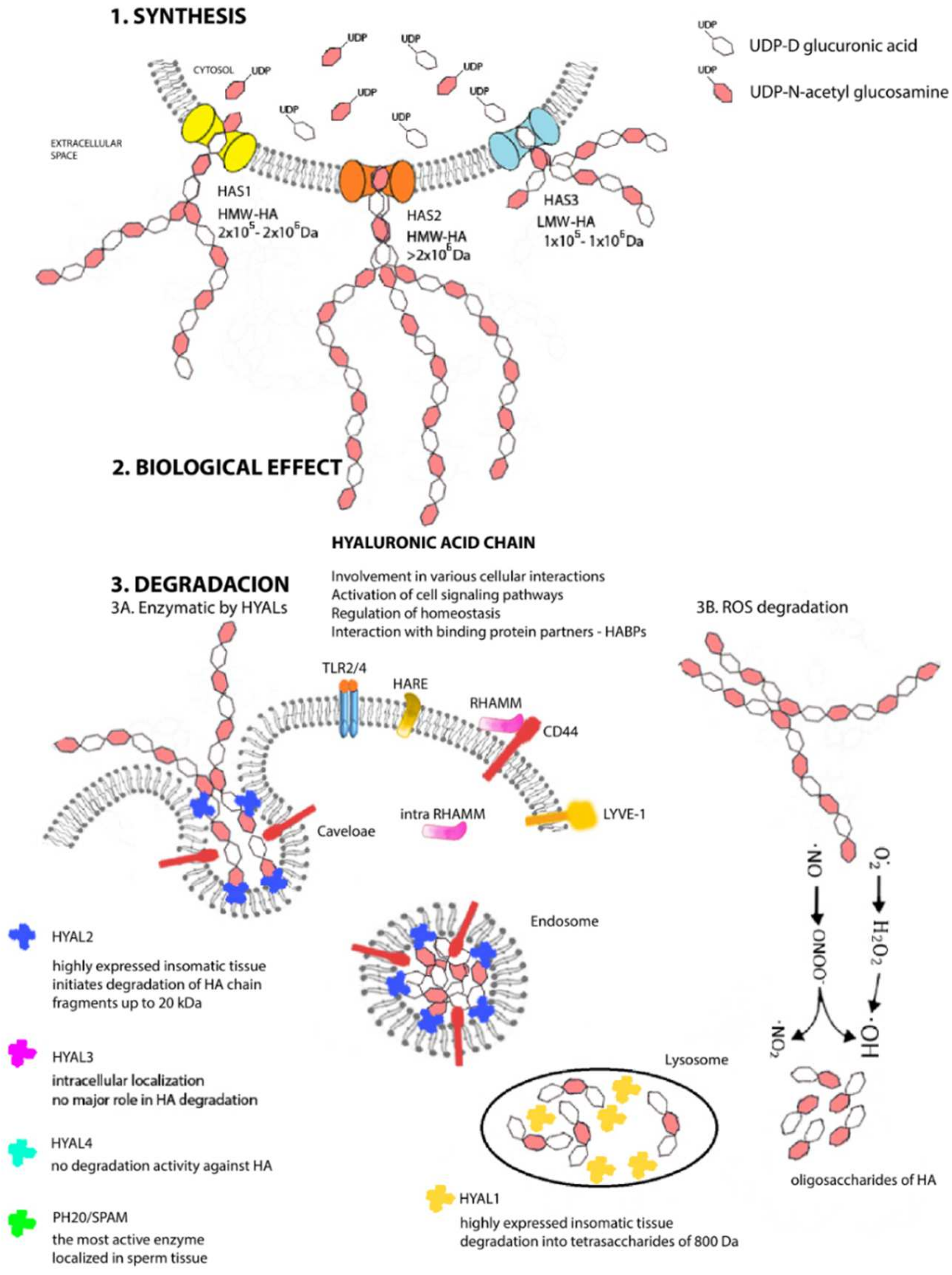
2. A Unique Metabolism from Beginning to End
This entry is adapted from the peer-reviewed paper 10.3390/pharmaceutics14040838
References
- Cowman, M.K. Hyaluronan and Hyaluronan Fragments. Adv. Carbohydr. Chem. Biochem. 2017, 74, 1–59.
- Laurent, T.C. Hyaluronan Research in Uppsala*. Upsala J. Med. Sci. 2007, 112, 123–142.
- Couchman, J.R.; Pataki, C.A. An Introduction to Proteoglycans and Their Localization. J. Histochem. Cytochem. 2012, 60, 885–897.
- Iozzo, R.V.; Schaefer, L. Proteoglycan Form and Function: A Comprehensive Nomenclature of Proteoglycans. Matrix Biol. 2015, 42, 11–55.
- Kakehi, K.; Kinoshita, M.; Yasueda, S. Hyaluronic Acid: Separation and Biological Implications. J. Chromatogr. B 2003, 797, 347–355.
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolář, J. Hyaluronic Acid (Hyaluronan): A Review. Vet. Med. 2008, 53, 397–411.
- Liao, Y.-H.; Jones, S.A.; Forbes, B.; Martin, G.P.; Brown, M.B. Hyaluronan: Pharmaceutical Characterization and Drug Delivery. Drug Deliv. 2005, 12, 327–342.
- Meyer, K.; Palmer, J.W. The Polysaccharide of the Vitreous Humor. J. Biol. Chem. 1934, 107, 629–634.
- Levene, P.A.; López-Suárez, J. Mucins and Mucoids. J. Biol. Chem. 1918, 36, 105–126.
- Weissmann, B.; Meyer, K. The Structure of Hyalobiuronic Acid and of Hyaluronic Acid from Umbilical Cord1,2. J. Am. Chem. Soc. 1954, 76, 1753–1757.
- Chen, W.Y.; Abatangelo, G. Functions of Hyaluronan in Wound Repair. Wound Repair Regen 1999, 7, 79–89.
- Hargittai, I.; Hargittai, M. Molecular Structure of Hyaluronan: An Introduction. Struct. Chem. 2008, 19, 697–717.
- Valcarcel, J.; Novoa-Carballal, R.; Pérez-Martín, R.I.; Reis, R.L.; Vázquez, J.A. Glycosaminoglycans from Marine Sources as Therapeutic Agents. Biotechnol. Adv. 2017, 35, 711–725.
- Sze, J.H.; Brownlie, J.C.; Love, C.A. Biotechnological Production of Hyaluronic Acid: A Mini Review. 3 Biotech 2016, 6, 67.
- Almond, A. Hyaluronan. Cell. Mol. Life Sci. 2007, 64, 1591–1596.
- Dechert, T.A.; Ducale, A.E.; Ward, S.I.; Yager, D.R. Hyaluronan in Human Acute and Chronic Dermal Wounds. Wound Repair Regen. 2006, 14, 252–258.
- Rah, M.J. A Review of Hyaluronan and Its Ophthalmic Applications. Optom. J. Am. Optom. Assoc. 2011, 82, 38–43.
- Khunmanee, S.; Jeong, Y.; Park, H. Crosslinking Method of Hyaluronic-Based Hydrogel for Biomedical Applications. J. Tissue Eng. 2017, 8, 2041731417726464.
- Jouon, N.; Rinaudo, M.; Milas, M.; Desbrières, J. Hydration of Hyaluronic Acid as a Function of the Counterion Type and Relative Humidity. Carbohydr. Polym. 1995, 26, 69–73.
- Nakamura, M.; Hikida, M.; Nakano, T.; Ito, S.; Hamano, T.; Kinoshita, S. Characterization of Water Retentive Properties of Hyaluronan. Cornea 1993, 12, 433–436.
- Huang, H.; Du, W.; Brekken, R.A. Extracellular Matrix Induction of Intracellular Reactive Oxygen Species. Antioxid. Redox Signal. 2017, 27, 774–784.
- Waddington, R.J.; Moseley, R.; Embery, G. Periodontal Disease Mechanisms: Reactive Oxygen Species: A Potential Role in the Pathogenesis of Periodontal Diseases. Oral Dis. 2000, 6, 138–151.
- Moseley, R.; Leaver, M.; Walker, M.; Waddington, R.J.; Parsons, D.; Chen, W.Y.J.; Embery, G. Comparison of the Antioxidant Properties of HYAFF®-11p75, AQUACEL® and Hyaluronan towards Reactive Oxygen Species in Vitro. Biomaterials 2002, 23, 2255–2264.
- DeAngelis, P.L. Hyaluronan Synthases: Fascinating Glycosyltransferases from Vertebrates, Bacterial Pathogens, and Algal Viruses. Cell. Mol. Life Sci. 1999, 56, 670–682.
- DeAngelis, P.L.; Papaconstantinou, J.; Weigel, P.H. Molecular Cloning, Identification, and Sequence of the Hyaluronan Synthase Gene from Group A Streptococcus Pyogenes. J. Biol. Chem. 1993, 268, 19181–19184.
- DeAngelis, P.L.; Papaconstantinou, J.; Weigel, P.H. Isolation of a Streptococcus Pyogenes Gene Locus That Directs Hyaluronan Biosynthesis in Acapsular Mutants and in Heterologous Bacteria. J. Biol. Chem. 1993, 268, 14568–14571.
- DeAngelis, P.L.; Jing, W.; Drake, R.R.; Achyuthan, A.M. Identification and Molecular Cloning of a Unique Hyaluronan Synthase from Pasteurella Multocida. J. Biol. Chem. 1998, 273, 8454–8458.
- Weigel, P.H. Functional Characteristics and Catalytic Mechanisms of the Bacterial Hyaluronan Synthases. IUBMB Life 2002, 54, 201–211.
- Weigel, P.H.; DeAngelis, P.L. Hyaluronan Synthases: A Decade-plus of Novel Glycosyltransferases. J. Biol. Chem. 2007, 282, 36777–36781.
- Weigel, P.H. Hyaluronan Synthase: The Mechanism of Initiation at the Reducing End and a Pendulum Model for Polysaccharide Translocation to the Cell Exterior. Int. J. Cell Biol. 2015, 2015, 367579.
- Siiskonen, H.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K. Hyaluronan Synthase 1: A Mysterious Enzyme with Unexpected Functions. Front. Immunol. 2015, 6, 43.
- Reitinger, S.; Müllegger, J.; Lepperdinger, G. Xenopus Kidney Hyaluronidase-1 (XKH1), a Novel Type of Membrane-Bound Hyaluronidase Solely Degrades Hyaluronan at Neutral PH. FEBS Lett. 2001, 505, 213–216.
- Tsepilov, R.N.; Beloded, A.V. Hyaluronic Acid-an “Old” Molecule with “New” Functions: Biosynthesis and Depolymerization of Hyaluronic Acid in Bacteria and Vertebrate Tissues Including during Carcinogenesis. Biochem. Mosc. 2015, 80, 1093–1108.
- Spicer, A.P.; McDonald, J.A. Characterization and Molecular Evolution of a Vertebrate Hyaluronan Synthase Gene Family. J. Biol. Chem. 1998, 273, 1923–1932.
- Fallacara, A.; Baldini, E.; Manfredini, S.; Vertuani, S. Hyaluronic Acid in the Third Millennium. Polymers 2018, 10, 701.
- Itano, N.; Kimata, K. Mammalian Hyaluronan Synthases. IUBMB Life 2002, 54, 195–199.
- Lee, J.Y.; Spicer, A.P. Hyaluronan: A Multifunctional, MegaDalton, Stealth Molecule. Curr. Opin. Cell Biol. 2000, 12, 581–586.
- Bai, K.-J.; Spicer, A.P.; Mascarenhas, M.M.; Yu, L.; Ochoa, C.D.; Garg, H.G.; Quinn, D.A. The Role of Hyaluronan Synthase 3 in Ventilator-Induced Lung Injury. Am. J. Respir. Crit. Care Med. 2005, 172, 92–98.
- Itano, N.; Sawai, T.; Yoshida, M.; Lenas, P.; Yamada, Y.; Imagawa, M.; Shinomura, T.; Hamaguchi, M.; Yoshida, Y.; Ohnuki, Y.; et al. Three Isoforms of Mammalian Hyaluronan Synthases Have Distinct Enzymatic Properties. J. Biol. Chem. 1999, 274, 25085–25092.
- Camenisch, T.D.; Spicer, A.P.; Brehm-Gibson, T.; Biesterfeldt, J.; Augustine, M.L.; Calabro, A.; Kubalak, S.; Klewer, S.E.; McDonald, J.A. Disruption of Hyaluronan Synthase-2 Abrogates Normal Cardiac Morphogenesis and Hyaluronan-Mediated Transformation of Epithelium to Mesenchyme. J. Clin. Investig. 2000, 106, 349–360.
- Tien, J.Y.L.; Spicer, A.P. Three Vertebrate Hyaluronan Synthases Are Expressed during Mouse Development in Distinct Spatial and Temporal Patterns. Dev. Dyn. 2005, 233, 130–141.
- Laurent, T.C.; Fraser, J.R. Hyaluronan. FASEB J. 1992, 6, 2397–2404.
- Vigetti, D.; Viola, M.; Karousou, E.; De Luca, G.; Passi, A. Metabolic Control of Hyaluronan Synthases. Matrix Biol. 2014, 35, 8–13.
- Vigetti, D.; Karousou, E.; Viola, M.; Passi, A. Analysis of Hyaluronan Synthase Activity. In Glycosaminoglycans: Chemistry and Biology; Balagurunathan, K., Nakato, H., Desai, U.R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 201–208. ISBN 978-1-4939-1714-3.
- Cowman, M.K.; Matsuoka, S. Experimental Approaches to Hyaluronan Structure. Carbohydr. Res. 2005, 340, 791–809.
- Tammi, R.H.; Passi, A.G.; Rilla, K.; Karousou, E.; Vigetti, D.; Makkonen, K.; Tammi, M.I. Transcriptional and Post-Translational Regulation of Hyaluronan Synthesis. FEBS J. 2011, 278, 1419–1428.
- Jacobson, A.; Brinck, J.; Briskin, M.J.; Spicer, A.P.; Heldin, P. Expression of Human Hyaluronan Synthases in Response to External Stimuli. Biochem. J. 2000, 348, 29–35.
- Tlapak-Simmons, V.L.; Baron, C.A.; Gotschall, R.; Haque, D.; Canfield, W.M.; Weigel, P.H. Hyaluronan Biosynthesis by Class I Streptococcal Hyaluronan Synthases Occurs at the Reducing End. J. Biol. Chem. 2005, 280, 13012–13018.
- Deen, A.J.; Rilla, K.; Oikari, S.; Kärnä, R.; Bart, G.; Häyrinen, J.; Bathina, A.R.; Ropponen, A.; Makkonen, K.; Tammi, R.H.; et al. Rab10-Mediated Endocytosis of the Hyaluronan Synthase HAS3 Regulates Hyaluronan Synthesis and Cell Adhesion to Collagen. J. Biol. Chem. 2014, 289, 8375–8389.
- Karousou, E.; Kamiryo, M.; Skandalis, S.S.; Ruusala, A.; Asteriou, T.; Passi, A.; Yamashita, H.; Hellman, U.; Heldin, C.-H.; Heldin, P. The Activity of Hyaluronan Synthase 2 Is Regulated by Dimerization and Ubiquitination. J. Biol. Chem. 2010, 285, 23647–23654.
- Bart, G.; Vico, N.O.; Hassinen, A.; Pujol, F.M.; Deen, A.J.; Ruusala, A.; Tammi, R.H.; Squire, A.; Heldin, P.; Kellokumpu, S.; et al. Fluorescence Resonance Energy Transfer (FRET) and Proximity Ligation Assays Reveal Functionally Relevant Homo-and Heteromeric Complexes among Hyaluronan Synthases HAS1, HAS2, and HAS3. J. Biol. Chem. 2015, 290, 11479–11490.
- Marcellin, E.; Steen, J.A.; Nielsen, L.K. Insight into Hyaluronic Acid Molecular Weight Control. Appl. Microbiol. Biotechnol. 2014, 98, 6947–6956.
- Ferrer, V.P.; de Mari, T.L.; Gremski, L.H.; Trevisan Silva, D.; da Silveira, R.B.; Gremski, W.; Chaim, O.M.; Senff-Ribeiro, A.; Nader, H.B.; Veiga, S.S. A Novel Hyaluronidase from Brown Spider (Loxosceles Intermedia) Venom (Dietrich’s Hyaluronidase): From Cloning to Functional Characterization. PLoS Negl. Trop. Dis. 2013, 7, e2206.
- Kogan, G.; Šoltés, L.; Stern, R.; Gemeiner, P. Hyaluronic Acid: A Natural Biopolymer with a Broad Range of Biomedical and Industrial Applications. Biotechnol. Lett. 2007, 29, 17–25.
- Takeo, S.; Fujise, M.; Akiyama, T.; Habuchi, H.; Itano, N.; Matsuo, T.; Aigaki, T.; Kimata, K.; Nakato, H. In Vivo Hyaluronan Synthesis upon Expression of the Mammalian Hyaluronan Synthase Gene in Drosophila. J. Biol. Chem. 2004, 279, 18920–18925.
- Shiedlin, A.; Bigelow, R.; Christopher, W.; Arbabi, S.; Yang, L.; Maier, R.V.; Wainwright, N.; Childs, A.; Miller, R.J. Evaluation of Hyaluronan from Different Sources: Streptococcus Zooepidemicus, Rooster Comb, Bovine Vitreous, and Human Umbilical Cord. Biomacromolecules 2004, 5, 2122–2127.
- Cowman, M.K.; Lee, H.-G.; Schwertfeger, K.L.; McCarthy, J.B.; Turley, E.A. The Content and Size of Hyaluronan in Biological Fluids and Tissues. Front. Immunol. 2015, 6, 261.
- Papakonstantinou, E.; Karakiulakis, G. The “sweet” and “Bitter” Involvement of Glycosaminoglycans in Lung Diseases: Pharmacotherapeutic Relevance. Br. J. Pharmacol. 2009, 157, 1111–1127.
- Laurent, C.; Johnson-Wells, G.; Hellström, S.; Engström-Laurent, A.; Wells, A.F. Localization of Hyaluronan in Various Muscular Tissues. Cell Tissue Res. 1991, 263, 201–205.
- Armstrong, S.E.; Bell, D.R. Relationship between Lymph and Tissue Hyaluronan in Skin and Skeletal Muscle. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H2485–H2494.
- George, J.; Stern, R. Serum Hyaluronan and Hyaluronidase: Very Early Markers of Toxic Liver Injury. Clin. Chim. Acta 2004, 348, 189–197.
- Toole, B.P. Hyaluronan and Its Binding Proteins, the Hyaladherins. Curr. Opin. Cell Biol. 1990, 2, 839–844.
- Allison, D.D.; Grande-Allen, K.J. Review. Hyaluronan: A Powerful Tissue Engineering Tool. Tissue Eng. 2006, 12, 2131–2140.
- Tiwari, S.; Bahadur, P. Modified Hyaluronic Acid Based Materials for Biomedical Applications. Int. J. Biol. Macromol. 2019, 121, 556–571.
- Pomin, V.H.; Mulloy, B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11, 27.
- Hardingham, T.E.; Muir, H. The Specific Interaction of Hyaluronic Acid with Cartilage Proteoglycans. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1972, 279, 401–405.
- Hascall, V.C.; Heinegård, D. Aggregation of Cartilage Proteoglycans I. The Role of Hyaluronic Acid. J. Biol. Chem. 1974, 249, 4232–4241.
- Laurent, U.B.G.; Reed, R.K. Turnover of Hyaluronan in the Tissues. Adv. Drug Deliv. Rev. 1991, 7, 237–256.
- Triggs-Raine, B.; Natowicz, M.R. Biology of Hyaluronan: Insights from Genetic Disorders of Hyaluronan Metabolism. World J. Biol. Chem. 2015, 6, 110–120.
- Buhren, B.A.; Schrumpf, H.; Hoff, N.-P.; Bölke, E.; Hilton, S.; Gerber, P.A. Hyaluronidase: From Clinical Applications to Molecular and Cellular Mechanisms. Eur. J. Med. Res. 2016, 21, 5.
- Stern, R.; Jedrzejas, M.J. The Hyaluronidases: Their Genomics, Structures, and Mechanisms of Action. Chem. Rev. 2006, 106, 818–839.
- Rigden, D.J.; Jedrzejas, M.J. Structures of Streptococcus Pneumoniae Hyaluronate Lyase in Complex with Chondroitin and Chondroitin Sulfate Disaccharides. Insights into Specificity and Mechanism of Action. J. Biol. Chem. 2003, 278, 50596–50606.
- Meyer, K. 11 Hyaluronidases. In The Enzymes; Boyer, P.D., Ed.; Academic Press: Cambridge, MA, USA, 1971; Volume 5, pp. 307–320.
- Jedrzejas, M.J. Structural and Functional Comparison of Polysaccharide-Degrading Enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 221–251.
- Khan, N.; Niazi, Z.R.; Rehman, F.U.; Akhtar, A.; Khan, M.M.; Khan, S.; Baloch, N.; Khan, S. Hyaluronidases: A Therapeutic Enzyme. Available online: https://www.ingentaconnect.com/content/ben/ppl/2018/00000025/00000007/art00010 (accessed on 2 April 2019).
- Stern, R. Devising a Pathway for Hyaluronan Catabolism: Are We There Yet? Glycobiology 2003, 13, 105R–115R.
- Kreil, G. Hyaluronidases-A Group of Neglected Enzymes. Protein Sci. 1995, 4, 1666–1669.
- Csóka, A.B.; Scherer, S.W.; Stern, R. Expression Analysis of Six Paralogous Human Hyaluronidase Genes Clustered on Chromosomes 3p21 and 7q31. Genomics 1999, 60, 356–361.
- Csóka, A.B.; Frost, G.I.; Wong, T.; Stern, R.; Csóka, T.B. Purification and Microsequencing of Hyaluronidase Isozymes from Human Urine. FEBS Lett. 1997, 417, 307–310.
- Frost, G.I.; Csóka, A.B.; Wong, T.; Stern, R.; Csóka, T.B. Purification, Cloning, and Expression of Human Plasma Hyaluronidase. Biochem. Biophys. Res. Commun. 1997, 236, 10–15.
- Frost, G.I.; Mohapatra, G.; Wong, T.M.; Csóka, A.B.; Gray, J.W.; Stern, R. HYAL1LUCA-1, a Candidate Tumor Suppressor Gene on Chromosome 3p21.3, Is Inactivated in Head and Neck Squamous Cell Carcinomas by Aberrant Splicing of Pre-MRNA. Oncogene 2000, 19, 870–877.
- Lepperdinger, G.; Müllegger, J.; Kreil, G. Hyal2-Less Active, but More Versatile? Matrix Biol. 2001, 20, 509–514.
- Rodén, L.; Campbell, P.; Fraser, J.R.; Laurent, T.C.; Pertoft, H.; Thompson, J.N. Enzymic Pathways of Hyaluronan Catabolism. Ciba Found. Symp. 1989, 143, 60–76; discussion 76–86, 281–285.
- Lepperdinger, G.; Strobl, B.; Kreil, G. HYAL2, a Human Gene Expressed in Many Cells, Encodes a Lysosomal Hyaluronidase with a Novel Type of Specificity. J. Biol. Chem. 1998, 273, 22466–22470.
- Vigetti, D.; Karousou, E.; Viola, M.; Deleonibus, S.; De Luca, G.; Passi, A. Hyaluronan: Biosynthesis and Signaling. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2452–2459.
- Stern, R. Hyaluronidases in Cancer Biology. Semin. Cancer Biol. 2008, 18, 275–280.
- Stern, R. Hyaluronan Catabolism: A New Metabolic Pathway. Eur. J. Cell Biol. 2004, 83, 317–325.
- Erickson, M.; Stern, R. Chain Gangs: New Aspects of Hyaluronan Metabolism. Biochem. Res. Int. 2011, 2012, e893947.
- Lin, Y.; Mahan, K.; Lathrop, W.F.; Myles, D.G.; Primakoff, P. A Hyaluronidase Activity of the Sperm Plasma Membrane Protein PH-20 Enables Sperm to Penetrate the Cumulus Cell Layer Surrounding the Egg. J. Cell Biol. 1994, 125, 1157–1163.
- Maciej-Hulme, M.L. New Insights into Human Hyaluronidase 4/Chondroitin Sulphate Hydrolase. Front. Cell Dev. Biol. 2021, 9, 1–7.
- Yamamoto, H.; Tobisawa, Y.; Inubushi, T.; Irie, F.; Ohyama, C.; Yamaguchi, Y. A Mammalian Homolog of the Zebrafish Transmembrane Protein 2 (TMEM2) Is the Long-Sought-after Cell-Surface Hyaluronidase. J. Biol. Chem. 2017, 292, 7304–7313.
- Stern, R.; Maibach, H.I. Hyaluronan in Skin: Aspects of Aging and Its Pharmacologic Modulation. Clin. Dermatol. 2008, 26, 106–122.
- Stern, R.; Kogan, G.; Jedrzejas, M.J.; Soltés, L. The Many Ways to Cleave Hyaluronan. Biotechnol. Adv. 2007, 25, 537–557.
- Monzon, M.E.; Fregien, N.; Schmid, N.; Falcon, N.S.; Campos, M.; Casalino-Matsuda, S.M.; Forteza, R.M. Reactive Oxygen Species and Hyaluronidase 2 Regulate Airway Epithelial Hyaluronan Fragmentation. J. Biol. Chem. 2010, 285, 26126–26134.
- Salwowska, N.M.; Bebenek, K.A.; Żądło, D.A.; Wcisło-Dziadecka, D.L. Physiochemical Properties and Application of Hyaluronic Acid: A Systematic Review. J. Cosmet. Derm. 2016, 15, 520–526.
- Heitzmann, E.; Thumm, D.; Baudouin, C. A Review of the Efficacy, Safety and Tolerability of Lacrycon® Eye Drops for the Treatment of Dry Eye Syndrome. J. Français D’Ophtalmologie 2019, 42, 642–654.
- López-Ruiz, E.; Jiménez, G.; Álvarez de Cienfuegos, L.; Antic, C.; Sabata, R.; Marchal, J.A.; Gálvez-Martín, P. Advances of Hyaluronic Acid in Stem Cell Therapy and Tissue Engineering, Including Current Clinical Trials. Eur. Cell. Mater. 2019, 37, 186–213.
- Huang, G.; Huang, H. Hyaluronic Acid-Based Biopharmaceutical Delivery and Tumor-Targeted Drug Delivery System. J. Control. Release 2018, 278, 122–126.
- Kim, J.H.; Moon, M.J.; Kim, D.Y.; Heo, S.H.; Jeong, Y.Y. Hyaluronic Acid-Based Nanomaterials for Cancer Therapy. Polymers 2018, 10, 1133.
