Infectious diseases are a burden for aquaculture. Antigen processing and presentation (APP) to the immune effector cells that fight pathogens is key in the adaptive immune response. At the core of the adaptive immunity that appeared in lower vertebrates during evolution are the variable genes encoding the major histocompatibility complex (MHC). MHC class I molecules mainly present peptides processed in the cytosol by the proteasome and transported to the cell surface of all cells through secretory compartments. Professional antigen-presenting cells (pAPC) also express MHC class II molecules, which normally present peptides processed from exogenous antigens through lysosomal pathways. Autophagy is an intracellular self-degradation process that is conserved in all eukaryotes and is induced by starvation to contribute to cellular homeostasis. Self-digestion during autophagy mainly occurs by the fusion of autophagosomes, which engulf portions of cytosol and fuse with lysosomes (macroautophagy) or assisted by chaperones (chaperone-mediated autophagy, CMA) that deliver proteins to lysosomes. Thus, during self-degradation, antigens can be processed to be presented by the MHC to immune effector cells, thus, linking autophagy to APP.
- antigen processing
- antigen-presenting cell
- bacteria
- chaperone-mediated autophagy
- LC3-Associated phagocytosis
- macroautophagy
- major histocompatibility complex MHC class I
- MHC class II
- vaccine
- virus
- teleost
1. Introduction
Antigen Processing and Presentation (APP) in Adaptive Immunity
2. Antigen Processing and Presentation in Teleost Fish
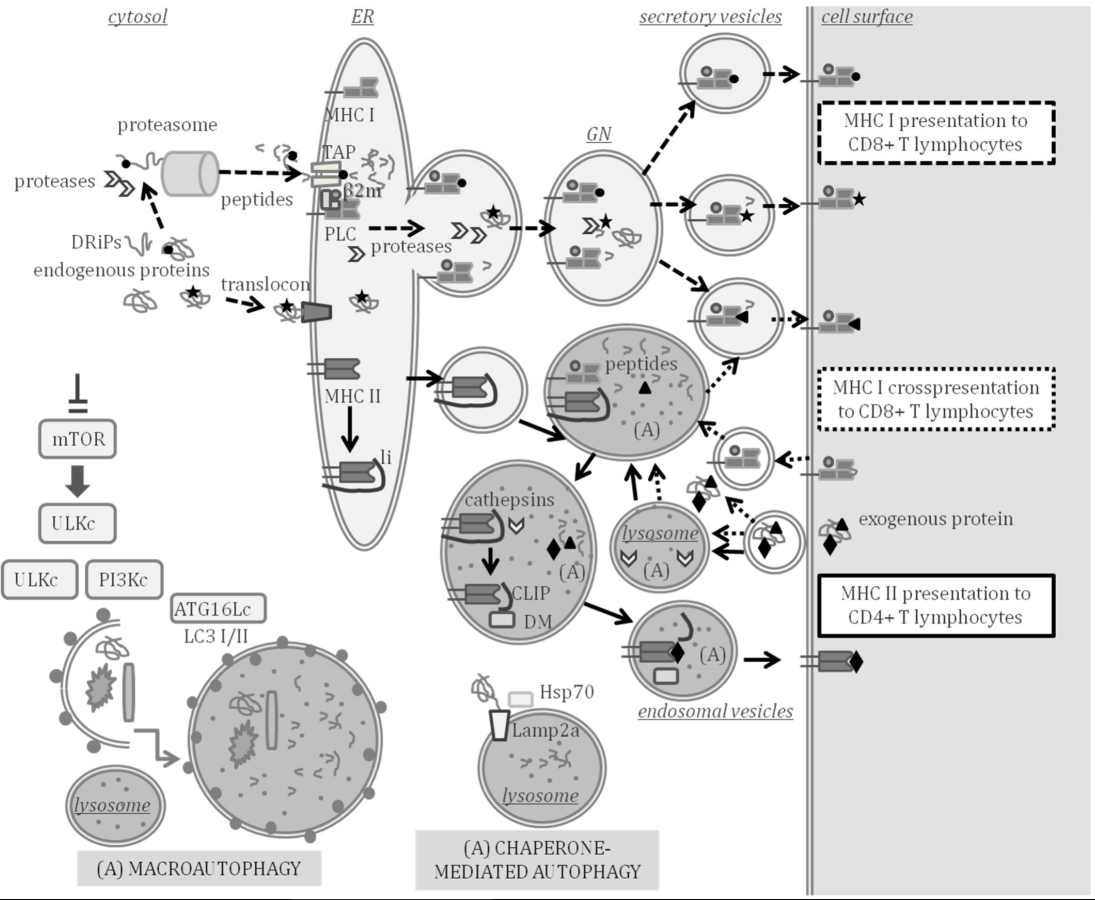
2.1. Antigen Processing and Presentation Genes Conserved in Teleost Fish
2.2. Antigen-Presenting Cells (APC) and Professional APC (pAPC) in Teleost Fish
2.3. Peptide Processing, Loading and Transport by MHC in Teleost Fish
3. Autophagy and Related LC3-Associated Phagocytosis in Teleost Fish
3.1. Chaperone-Mediated Autophagy in Teleost Fish
3.2. Macroautophagy in Teleost Fish
3.3. LC3-Associated Phagocytosis in Teleost Fish
This entry is adapted from the peer-reviewed paper 10.3390/ijms23094899
References
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836.
- Münz, C. The macroautophagy machinery in MHC restricted antigen presentation. Front. Immunol. 2021, 12, 1–7.
- Münz, C. Non-canonical functions of autophagy proteins in immunity and infection. Mol. Asp. Med. 2021, 82, 100987.
- Rock, K.L.; Reits, E.; Neefjes, J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 2016, 37, 724–737.
- Wilson, A.B. MHC and adaptive immunity in teleost fishes. Immunogenetics 2017, 69, 521–528.
- Vallejo, A.N.; Miller, N.W.; Clem, L.W. Phylogeny of immune recognition: Role of alloantigens in antigen presentation in channel catfish immune responses. Immunology 1991, 74, 165–168.
- Vallejo, A.N.; Miller, N.W.; Harvey, N.E.; Cuchens, M.A.; William Clem, L. Cellular pathway(s) of antigen processing and presentation in fish APC: Endosomal involvement and cell-free antigen presentation. Dev. Immunol. 1992, 3, 51–65.
- Yamaguchi, T.; Dijkstra, J.M. Major histocompatibility complex (MHC) genes and disease resistance in fish. Cells 2019, 8, 378.
- Radwan, J.; Babik, W.; Kaufman, J.; Lenz, T.L.; Winternitz, J. Advances in the evolutionary understanding of MHC polymorphism. Trends Genet. 2020, 36, 298–311.
- Kaufman, J. Unfinished business: Evolution of the MHC and the adaptive immune system of jawed vertebrates. Annu. Rev. Immunol. 2018, 36, 383–409.
- Nonaka, M.; Naruse, K.; Matsuo, M.; Shima, A. Comparative genomics of medaka: The major histocompatibility complex (MHC). Mar. Biotechnol. 2001, 3, S141–S144.
- Kim, D.-H.; Lee, S.-M.; Hong, B.-Y.; Kim, Y.-T.; Choi, T.-J. Cloning and sequence analysis of cDNA for the proteasome activator PA28-β subunit of flounder (Paralichthys olivaceus). Mol. Immunol. 2003, 40, 611–616.
- Liu, G.; Zheng, W.; Chen, X. Molecular cloning of proteasome activator PA28-β subunit of large yellow croaker (Pseudosciana crocea) and its coordinated up-regulation with MHC class I α-chain and β2-microglobulin in poly I:C-treated fish. Mol. Immunol. 2007, 44, 1190–1197.
- Kasthuri, S.R.; Umasuthan, N.; Whang, I.; Kim, E.; Park, H.-C.; Lee, J. Genomic structural characterization and transcriptional expression analysis of proteasome activator PA28α and PA28β subunits from Oplegnathus fasciatus. Fish Shellfish Immunol. 2013, 35, 1224–1234.
- Pinto, R.D.; Randelli, E.; Buonocore, F.; Pereira, P.J.B.; dos Santos, N.M.S. Molecular cloning and characterization of sea bass (Dicentrarchus labrax, L.) MHC class I heavy chain and β2-microglobulin. Dev. Comp. Immunol. 2013, 39, 234–254.
- Pinto, R.D.; Moreira, A.R.; Pereira, P.J.B.; dos Santos, N.M.S. Two thioredoxin-superfamily members from sea bass (Dicentrarchus labrax, L.): Characterization of PDI (PDIA1) and ERp57 (PDIA3). Fish Shellfish Immunol. 2013, 35, 1163–1175.
- Sever, L.; Bols, N.C.; Dixon, B. The cloning and inducible expression of the rainbow trout ERp57 gene. Fish Shellfish Immunol. 2013, 34, 410–419.
- Sever, L.; Vo, N.T.K.; Bols, N.C.; Dixon, B. Expression of tapasin in rainbow trout tissues and cell lines and up regulation in a monocyte/macrophage cell line (RTS11) by a viral mimic and viral infection. Dev. Comp. Immunol. 2014, 44, 86–93.
- Wilson, W.H.; Gilg, I.C.; Moniruzzaman, M.; Field, E.K.; Koren, S.; Lecleir, G.R.; Martínez-Martínez, J.; Poulton, N.J.; Swan, B.K.; Stepanauskas, R.; et al. Genomic exploration of individual giant ocean viruses. ISME J. 2017, 11, 1736–1745.
- Nonaka, M.I.; Aizawa, K.; Mitani, H.; Bannai, H.P.; Nonaka, M. Retained orthologous relationships of the MHC class I genes during euteleost evolution. Mol. Biol. Evol. 2011, 28, 3099–3112.
- Chen, J.; Wang, L.; Huang, J.; Li, X.; Guan, L.; Wang, Q.; Yang, M.; Qin, Q. Functional analysis of a novel MHC-Iα genotype in orange-spotted grouper: Effects on Singapore grouper iridovirus (SGIV) replication and apoptosis. Fish Shellfish Immunol. 2022, 121, 487–497.
- Hashimoto, K.; Nakanishi, T.; Kurosawa, Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc. Natl. Acad. Sci. USA 1990, 87, 6863–6867.
- McConnell, S.C.; Hernández, K.M.; Wcisel, D.J.; Kettleborough, R.N.; Stemple, D.L.; Yoder, J.A.; Andrade, J.; de Jong, J.L.O. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc. Natl. Acad. Sci. USA 2016, 113, E5014–E5023.
- Grimholt, U. Whole genome duplications have provided teleosts with many roads to peptide loaded MHC class I molecules. BMC Evol. Biol. 2018, 18, 1–14.
- Dirscherl, H.; Yoder, J.A. A nonclassical MHC class I U lineage locus in zebrafish with a null haplotypic variant. Immunogenetics 2015, 67, 501–513.
- Grimholt, U.; Fosse, J.H.; Sundaram, A.Y.M. Selective stimulation of duplicated Atlantic salmon MHC pathway genes by interferon-gamma. Front. Immunol. 2020, 11, 571650–571665.
- Cuesta, A.; Ángeles Esteban, M.; Meseguer, J. Cloning, distribution and up-regulation of the teleost fish MHC class II alpha suggests a role for granulocytes as antigen-presenting cells. Mol. Immunol. 2006, 43, 1275–1285.
- Pilstrom, L.; Warr, G.W.; Stromberg, S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fish. Sci. 2005, 71, 961–971.
- Haase, D.; Roth, O.; Kalbe, M.; Schmiedeskamp, G.; Scharsack, J.P.; Rosenstiel, P.; Reusch, T.B.H. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: Evidence from deep transcriptome sequencing. Biol. Lett. 2013, 9, 20130044.
- Dijkstra, J.M.; Yamaguchi, T. Ancient features of the MHC class II presentation pathway, and a model for the possible origin of MHC molecules. Immunogenetics 2019, 71, 233–249.
- Dijkstra, J.M.; Grimholt, U.; Leong, J.; Koop, B.F.; Hashimoto, K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol. Biol. 2013, 13, 260.
- Fujiki, K. Alternate forms of MHC class II-associated invariant chain are not produced by alternative splicing in rainbow trout (Oncorhynchus mykiss) but are encoded by separate genes. Dev. Comp. Immunol. 2003, 27, 377–391.
- Semple, S.L.; Heath, G.; Christie, D.; Braunstein, M.; Kales, S.C.; Dixon, B. Immune stimulation of rainbow trout reveals divergent regulation of MH class II-associated invariant chain isoforms. Immunogenetics 2019, 71, 407–420.
- Li, M.; Li, Q.; Yang, Z.; Hu, G.; Li, T.; Chen, X.; Ao, J. Identification of cathepsin B from large yellow croaker (Pseudosciaena crocea) and its role in the processing of MHC class II-associated invariant chain. Dev. Comp. Immunol. 2014, 45, 313–320.
- Chen, L.; Zhang, M.; Sun, L. Identification and expressional analysis of two cathepsins from half-smooth tongue sole (Cynoglossus semilaevis). Fish. Shellfish Immunol. 2011, 31, 1270–1277.
- Li, Q.; Ao, J.; Mu, Y.; Yang, Z.; Li, T.; Zhang, X.; Chen, X. Cathepsin S, but not cathepsin L, participates in the MHC class II-associated invariant chain processing in large yellow croaker (Larimichthys crocea). Fish. Shellfish Immunol. 2015, 47, 743–750.
- Sun, Y.; Xu, T.; Wang, J.; Cheng, Y.; Wang, R. Sequence and expression analysis of cathepsin S gene in the miiuy croaker Miichthys miiuy. Fish. Physiol. Biochem. 2011, 37, 761–765.
- Che, R.; Wang, R.; Xu, T. Comparative genomic of the teleost cathepsin B and H and involvement in bacterial induced immunity of miiuy croaker. Fish. Shellfish Immunol. 2014, 41, 163–171.
- Dong, X.; Ye, Z.; Song, L.; Su, B.; Zhao, H.; Peatman, E.; Li, C. Expression profile analysis of two cathepsin S in channel catfish (Ictalurus punctatus) mucosal tissues following bacterial challenge. Fish. Shellfish Immunol. 2016, 48, 112–118.
- Shen, Y.; Zhang, H.; Zhou, Y.; Sun, Y.; Yang, H.; Cao, Z.; Qin, Q.; Liu, C.; Guo, W. Functional characterization of cathepsin B and its role in the antimicrobial immune responses in golden pompano (Trachinotus ovatus). Dev. Comp. Immunol. 2021, 123, 104128.
- Criscitiello, M.F.; Ohta, Y.; Graham, M.D.; Eubanks, J.O.; Chen, P.L.; Flajnik, M.F. Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev. Comp. Immunol. 2012, 36, 521–533.
- Dijkstra, J.M.; Köllner, B.; Aoyagi, K.; Sawamoto, Y.; Kuroda, A.; Ototake, M.; Nakanishi, T.; Fischer, U. The rainbow trout classical MHC class I molecule Onmy-UBA*501 is expressed in similar cell types as mammalian classical MHC class I molecules. Fish. Shellfish Immunol. 2003, 14, 1–23.
- Chang, Y.T.; Kai, Y.H.; Chi, S.C.; Song, Y.L. Cytotoxic CD8α+ leucocytes have heterogeneous features in antigen recognition and class I MHC restriction in grouper. Fish. Shellfish Immunol. 2011, 30, 1283–1293.
- Kanako, L. Lewis; Natasha, Del Cid; D.T. Perspectives on antigen presenting cells in zebrafish. Dev. Comp. Immunol. 2014, 46, 63–73.
- Iliev, D.B.; Thim, H.; Lagos, L.; Olsen, R.; Jørgensen, J.B. Homing of antigen-presenting cells in head kidney and spleen—salmon head kidney hosts diverse APC types. Front. Immunol. 2013, 4, 137.
- Ronza, P.; Álvarez-Dios, J.A.; Robledo, D.; Losada, A.P.; Romero, R.; Bermúdez, R.; Pardo, B.G.; Martínez, P.; Quiroga, M.I. Blood transcriptomics of turbot Scophthalmus maximus: A tool for health monitoring and disease studies. Animals 2021, 11, 1296.
- Pereiro, P.; Romero, A.; Díaz-Rosales, P.; Estepa, A.; Figueras, A.; Novoa, B. Nucleated teleost erythrocytes play an Nk-lysin- and autophagy-dependent role in antiviral immunity. Front. Immunol. 2017, 8, 1458.
- Lugo-Villarino, G.; Balla, K.M.; Stachura, D.L.; Bañuelos, K.; Werneck, M.B.F.; Traver, D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 15850–15855.
- Shao, T.; Zhu, L.-Y.; Nie, L.; Shi, W.; Dong, W.-R.; Xiang, L.-X.; Shao, J.-Z. Characterization of surface phenotypic molecules of teleost dendritic cells. Dev. Comp. Immunol. 2015, 49, 38–43.
- Zoccola, E.; Delamare-Deboutteville, J.; Barnes, A.C. Identification of barramundi (Lates calcarifer) DC-SCRIPT, a specific molecular marker for dendritic cells in fish. PLoS ONE 2015, 10, e0132687.
- Soleto, I.; Fischer, U.; Tafalla, C.; Granja, A.G. Identification of a potential common ancestor for mammalian cross-presenting dendritic cells in teleost respiratory surfaces. Front. Immunol. 2018, 9, 1–13.
- Soleto, I.; Granja, A.G.; Simón, R.; Morel, E.; Díaz-Rosales, P.; Tafalla, C. Identification of CD8α+ dendritic cells in rainbow trout (Oncorhynchus mykiss) intestine. Fish. Shellfish Immunol. 2019, 89, 309–318.
- Kato, G.; Miyazawa, H.; Nakayama, Y.; Ikari, Y.; Kondo, H.; Yamaguchi, T.; Sano, M.; Fischer, U. A novel antigen-sampling cell in the teleost gill epithelium with the potential for direct antigen presentation in mucosal tissue. Front. Immunol. 2018, 9, 2116–2128.
- Hu, Y.; Li, A.; Xu, Y.; Jiang, B.; Lu, G.; Luo, X. Transcriptomic variation of locally-infected skin of Epinephelus coioides reveals the mucosal immune mechanism against Cryptocaryon irritans. Fish. Shellfish Immunol. 2017, 66, 398–410.
- Wang, H.; Chen, X.; Li, S.; Zhou, C.; Xu, L.; Wu, Z.; Chen, X. Identification and expression analysis of Langerhans cells marker Langerin/CD207 in grass carp, Ctenopharyngodon idella. Gene 2021, 768, 145315.
- Wu, L.; Qin, Z.; Liu, H.; Lin, L.; Ye, J.; Li, J. Recent advances on phagocytic B cells in teleost fish. Front. Immunol. 2020, 11, 824–833.
- Miller, N.; Wilson, M.; Bfuflten, E.; Stuge, T.; Warr, G.; Ciem, W. Functional and molecular characterization of teleost leukocytes. Immunol. Rev. 1998, 166, 187–197.
- Abós, B.; Castro, R.; González Granja, A.; Havixbeck, J.J.; Barreda, D.R.; Tafalla, C. Early activation of teleost B cells in response to rhabdovirus infection. J. Virol. 2015, 89, 1768–1780.
- Castro, R.; Abós, B.; González, L.; Granja, A.G.; Tafalla, C. Expansion and differentiation of IgM+ B cells in the rainbow trout peritoneal cavity in response to different antigens. Dev. Comp. Immunol. 2017, 70, 119–127.
- Sunyer, J.O. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets 2012, 12, 200–212.
- Rougeot, J.; Torraca, V.; Zakrzewska, A.; Kanwal, Z.; Jansen, H.J.; Sommer, F.; Spaink, H.P.; Meijer, A.H. RNAseq profiling of leukocyte populations in zebrafish larvae reveals a cxcl11 chemokine gene as a marker of macrophage polarization during mycobacterial infection. Front. Immunol. 2019, 10, 832.
- Sever, L.; Vo, N.T.K.; Bols, N.C.; Dixon, B. Tapasin’s protein interactions in the rainbow trout peptide-loading complex. Dev. Comp. Immunol. 2018, 81, 262–270.
- Cui, X.; Ji, C.; Cao, X.; Fu, Z.; Zhang, S.; Guo, X. Molecular and biological characterization of interferon-γ-inducible-lysosomal thiol reductase gene in zebrafish (Danio rerio). Fish. Shellfish Immunol. 2012, 33, 1133–1138.
- Zheng, W.; Chen, X. Cloning and expression analysis of interferon-γ-inducible-lysosomal thiol reductase gene in large yellow croaker (Pseudosciaena crocea). Mol. Immunol. 2006, 43, 2135–2141.
- Song, J.; Liu, H.; Ma, L.; Gao, C.; Zhang, S. Molecular cloning, expression and functional characterization of interferon-γ-inducible lysosomal thiol reductase (GILT) gene from mandarin fish (Siniperca chuatsi). Fish. Shellfish Immunol. 2014, 38, 275–281.
- Gomes, L.C.; Dikic, I. Autophagy in antimicrobial immunity. Mol. Cell 2014, 54, 224–233.
- Herpin, A.; Lescat, L.; Bobe, J.; Jenny, A.; Seiliez, I. Lighting chaperone-mediated autophagy (CMA) evolution with an ancient LAMP: The existence of a functional CMA activity in fish. Autophagy 2020, 16, 1918–1920.
- Lescat, L.; Véeron, V.; Mourot, B.; Péron, S.; Chenais, N.; Dias, K.; Riera-Heredia, N.; Beaumatin, F.; Pinel, K.; Priault, M.; et al. Chaperone-mediated autophagy in the light of evolution: Insight from fish. Mol. Biol. Evol. 2020, 37, 2887–2899.
- Lescat, L.; Herpin, A.; Mourot, B.; Véron, V.; Guiguen, Y.; Bobe, J.; Seiliez, I. CMA restricted to mammals and birds: Myth or reality? Autophagy 2018, 14, 1267–1270.
- Yabu, T.; Imamura, S.; Mohammed, M.S.; Touhata, K.; Minami, T.; Terayama, M.; Yamashita, M. Differential gene expression of HSC70/HSP70 in yellowtail cells in response to chaperone-mediated autophagy. FEBS J. 2011, 278, 673–685.
- Dubińska-Magiera, M.; Niedbalska-Tarnowska, J.; Migocka-Patrzałek, M.; Posyniak, E.; Daczewska, M. Characterization of Hspb8 in Zebrafish. Cells 2020, 9, 1562.
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2021, 17, 1–382.
- Pant, D.C.; Nazarko, T.Y. Selective autophagy: The rise of the zebrafish model. Autophagy 2021, 17, 3297–3305.
- Muñoz-Sánchez, S.; van der Vaart, M.; Meijer, A.H. Autophagy and Lc3-associated phagocytosis in zebrafish models of bacterial infections. Cells 2020, 9, 2372.
- Valionyte, E.; Yang, Y.; Griffiths, S.A.; Bone, A.T.; Barrow, E.R.; Sharma, V.; Lu, B.; Luo, S. The caspase-6–p62 axis modulates p62 droplets based autophagy in a dominant-negative manner. Cell Death Differ. 2021.
- Dong, G.; Zhang, Z.; Duan, K.; Shi, W.; Huang, R.; Wang, B.; Luo, L.; Zhang, Y.; Ruan, H.; Huang, H. Beclin 1 deficiency causes hepatic cell apoptosis via endoplasmic reticulum stress in zebrafish larvae. FEBS Lett. 2020, 594, 1155–1165.
- Mawed, S.A.; He, Y.; Zhang, J.; Mei, J. Strategy of hepatic metabolic defects induced by beclin1 heterozygosity in adult zebrafish. Int. J. Mol. Sci. 2020, 21, 1533.
- Masud, S.; Prajsnar, T.K.; Torraca, V.; Lamers, G.E.M.; Benning, M.; Van Der Vaart, M.; Meijer, A.H. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy 2019, 15, 796–812.
- Prajsnar, T.K.; Serba, J.J.; Dekker, B.M.; Gibson, J.F.; Masud, S.; Fleming, A.; Johnston, S.A.; Renshaw, S.A.; Meijer, A.H. The autophagic response to Staphylococcus aureus provides an intracellular niche in neutrophils. Autophagy 2021, 17, 888–902.
- Gibson, J.F.; Prajsnar, T.K.; Hill, C.J.; Tooke, A.K.; Serba, J.J.; Tonge, R.D.; Foster, S.J.; Grierson, A.J.; Ingham, P.W.; Renshaw, S.A.; et al. Neutrophils use selective autophagy receptor Sqstm1/p62 to target Staphylococcus aureus for degradation in vivo in zebrafish. Autophagy 2021, 17, 1448–1457.
