Erythroid-related nuclear factor 2 (NRF2) and the antioxidant-responsive-elements (ARE) signaling pathway are the master regulators of cell antioxidant defenses, playing a key role in maintaining cellular homeostasis, a scenario in which proper mitochondrial function is essential. Increasing evidence indicates that the regular practice of physical exercise increases cellular antioxidant defenses by activating NRF2 signaling.
- NRF2
- oxidative stress
- exercise
- brain
- tetrahydrobiopterin
- neopterin
- epigenetics
1. Role of BH4 on NRF2/ARE Pathway Activated by Physical Exercise
2. Epigenetics as a Key Player in NRF2 Upregulation Induced by Physical Exercise
2.1. DNA Methylation
2.2. Histone Modifications
2.3. Post-Transcriptional Regulation
| Physical Exercise | LncRNA | Reported Effect | References |
|---|---|---|---|
| Swimming | CPhar | Prevention of myocardial ischemia-reperfusion injury and cardiac dysfunction | [63] |
| Swimming | Mhrt779 | Heart antihypertrophic effect | [64] |
| Treadmill | MSTRG.2625 MSTRG.1557 MSTRG.691 MSTRG.7497 |
Promotion of osteogenic differentiation | [65] |
| Treadmill | CYTOR | Regulation of fast-twitch myogenesis in aging | [66] |
| Aerobic exercise (single jump rope, double jump rope, round-trip running, and gymnastics) | MALAT1 | Improvement of endothelial dysfunction | [67] |
| Swimming | LOC102633466 LOC102637865 LOC102638670 |
Improved motor performance | [68] |
| Treadmill | TUG1 | Reduction of hippocampal neuronal apoptosis | [69] |
| Treadmill | Neat1 Meg3 Malat1 Kcnq1ot1 |
Possible involvement in insulin resistance and glucose homeostasis pathways | [70] |
| Running wheels | SNHG14 | Improvement of cognitive disorder and inflammation | [71] |
LncRNA studies provide new insights into the regulation of beneficial exercise-induced effects, but despite NRF2 having a central role in these effects, studies demonstrating the involvement of lncRNA in the regulation of exercise-induced NRF2 expression are scarce. Following aerobic exercise, miR-340-5p has been shown to play a role in the post-transcriptional regulation of NRF2 expression in mouse skeletal muscles [72].
3. Effects of BH4 on Epigenetic Modulation Induced by Physical Exercise
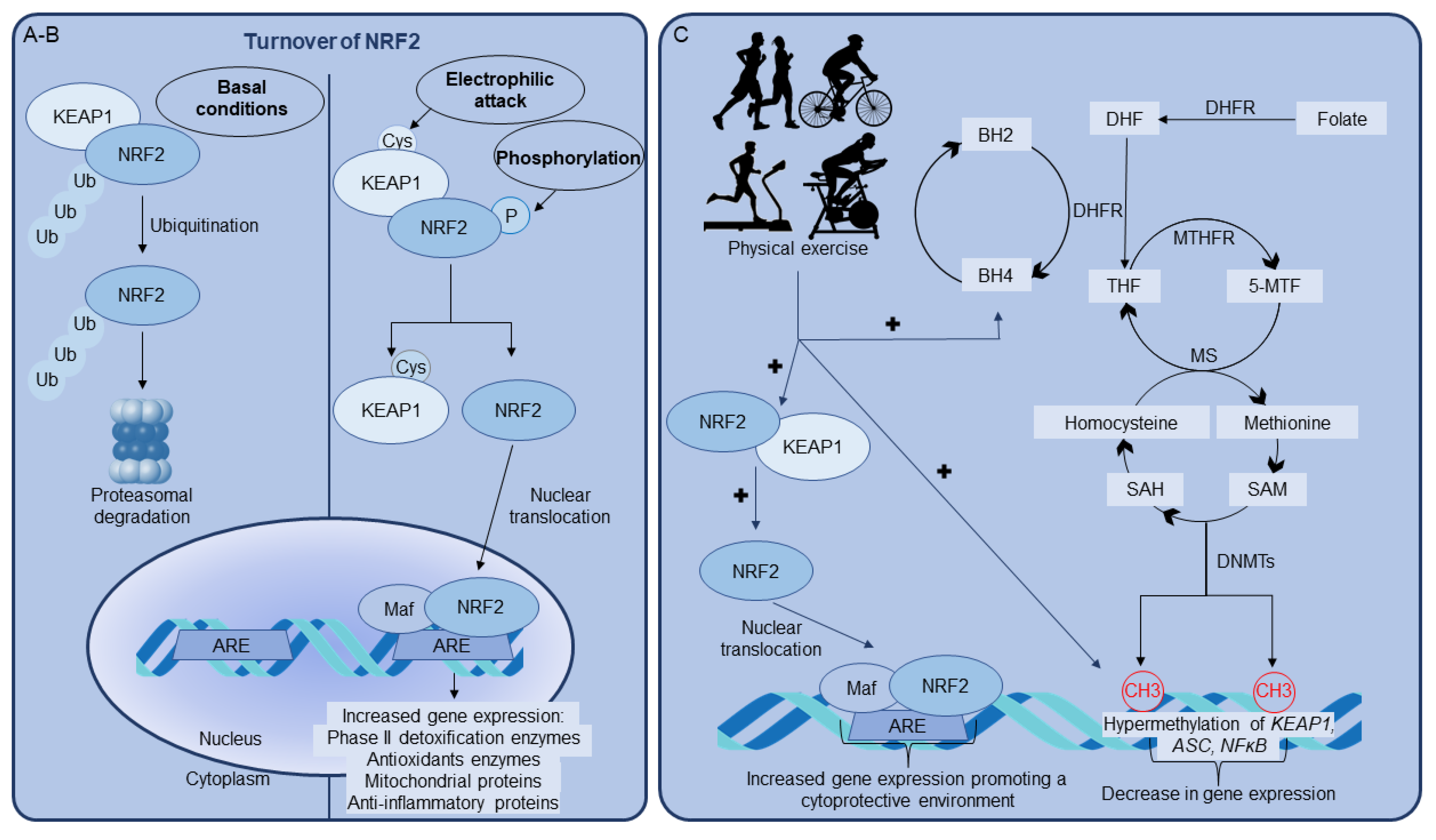
4. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/antiox11050826
References
- Thony, B.; Auerbach, G.; Blau, N.; Thöny, B.; Auerbach, G.; Blau, N.; Thöny, B.; Auerbach, G.; Blau, N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 2000, 347 Pt 1, 1–16.
- Werner, E.R.E.R.; Blau, N.; Thöny, B. Tetrahydrobiopterin: Biochemistry and pathophysiology. Biochem. J. 2011, 438, 397–414.
- Werner, E.R.; Werner-Felmayer, G.; Fuchs, D.; Hausen, A.; Reibnegger, G.; Yim, J.J.; Pfleiderer, W.; Wachter, H. Tetrahydrobiopterin biosynthetic activities in human macrophages, fibroblasts, THP-1, and T 24 cells. GTP-cyclohydrolase I is stimulated by interferon-gamma, and 6-pyruvoyl tetrahydropterin synthase and sepiapterin reductase are constitutively present. J. Biol. Chem. 1990, 265, 3189–3192.
- Ghisoni, K.; de Paula Martins, R.; Barbeito, L.; Latini, A. Neopterin as a potential cytoprotective brain molecule. J. Psychiatr. Res. 2015, 71, 134–139.
- Cronin, S.J.F.; Seehus, C.; Weidinger, A.; Talbot, S.; Reissig, S.; Seifert, M.; Pierson, Y.; McNeill, E.; Longhi, M.S.; Turnes, B.L.; et al. The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature 2018, 563, 564–568.
- Choi, H.J.; Lee, S.Y.; Cho, Y.; No, H.; Kim, S.W.; Hwang, O. Tetrahydrobiopterin causes mitochondrial dysfunction in dopaminergic cells: Implications for Parkinson’s disease. Neurochem. Int. 2006, 48, 255–262.
- Bailey, J.; Shaw, A.; Fischer, R.; Ryan, B.J.; Kessler, B.M.; McCullagh, J.; Wade-Martins, R.; Channon, K.M.; Crabtree, M.J. A novel role for endothelial tetrahydrobiopterin in mitochondrial redox balance. Free Radic. Biol. Med. 2017, 104, 214–225.
- Ghisoni, K.; Latini, A. Kuehne LK, Reiber H, Bechter K, Hagberg L, Fuchs D., Cerebrospinal fluid neopterin is brain-derived and not associated with blood-CSF barrier dysfunction in non-inflammatory affective and schizophrenic spectrum disorders. Letter to the Editor. J. Psychiatr. Res. 2015, 63, 141–142.
- Martins, R.D.P.; Ghisoni, K.; Lim, C.K.; Aguiar, A.S.; Guillemin, G.J.; Latini, A. Neopterin preconditioning prevents inflammasome activation in mammalian astrocytes. Free Radic. Biol. Med. 2018, 115, 371–382.
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of proIL-β. Mol. Cell 2002, 10, 417–426.
- Ghisoni, K.; Aguiar, A.S.; de Oliveira, P.A.; Matheus, F.C.; Gabach, L.; Perez, M.; Carlini, V.P.; Barbeito, L.; Mongeau, R.; Lanfumey, L.; et al. Neopterin acts as an endogenous cognitive enhancer. Brain. Behav. Immun. 2016, 56, 156–164.
- Tutakhail, A.; Nazary, Q.A.; Lebsir, D.; Kerdine-Romer, S.; Coudore, F. Induction of brain Nrf2-HO-1 pathway and antinociception after different physical training paradigms in mice. Life Sci. 2018, 209, 149–156.
- Aguiar, A.S.; Duzzioni, M.; Remor, A.P.; Tristão, F.S.M.; Matheus, F.C.; Raisman-Vozari, R.; Latini, A.; Prediger, R.D. Moderate-intensity physical exercise protects against experimental 6-hydroxydopamine-induced hemiparkinsonism through Nrf2-antioxidant response element pathway. Neurochem. Res. 2016, 41, 64–72.
- da Luz Scheffer, D.; Ghisoni, K.; Aguiar, A.S.; Latini, A. Moderate running exercise prevents excessive immune system activation. Physiol. Behav. 2019, 204, 248–255.
- McNeill, E.; Crabtree, M.J.; Sahgal, N.; Patel, J.; Chuaiphichai, S.; Iqbal, A.J.; Hale, A.B.; Greaves, D.R.; Channon, K.M. Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation. Free Radic. Biol. Med. 2015, 79, 206–216.
- Lindsay, A.; Costello, J.T. Realising the Potential of Urine and Saliva as Diagnostic Tools in Sport and Exercise Medicine. Sport. Med. 2017, 47, 11–31.
- Gieseg, S.; Baxter-Parker, G.; Lindsay, A. Neopterin, Inflammation, and Oxidative Stress: What Could We Be Missing? Antioxidants 2018, 7, 80.
- HASHIMOTO, R.; NAGATSU, T.; OHTA, T.; MIZUTANI, M.; OMURA, I. Changes in the Concentrations of Tetrahydrobiopterin, the Cofactor of Tyrosine Hydroxylase, in Blood under Physical Stress and in Depression. Ann. N. Y. Acad. Sci. 2004, 1018, 378–386.
- Mizutani, M.; Hashimoto, R.; Ohta, T.; Nakazawa, K.; Nagatsu, T. The Effect of Exercise on Plasma Biopterin Levels. Neuropsychobiology 1994, 29, 53–56.
- Finsterer, J. Biomarkers of peripheral muscle fatigue during exercise. BMC Musculoskelet. Disord. 2012, 13, 218.
- Sprenger, H.; Jacobs, C.; Nain, M.; Gressner, A.M.; Prinz, H.; Wesemann, W.; Gemsa, D. Enhanced release of cytokines, interleukin-2 receptors, and neopterin after long-distance running. Clin. Immunol. Immunopathol. 1992, 63, 188–195.
- Baxter-Parker, G.; Chu, A.; Petocz, P.; Samman, S.; Gieseg, S.P. Simultaneous analysis of neopterin, kynurenine and tryptophan by amine-HPLC shows minor oxidative stress from short-term exhaustion exercise. Pteridines 2019, 30, 21–32.
- Strasser, B.; Geiger, D.; Schauer, M.; Gatterer, H.; Burtscher, M.; Fuchs, D. Effects of Exhaustive Aerobic Exercise on Tryptophan-Kynurenine Metabolism in Trained Athletes. PLoS ONE 2016, 11, e0153617.
- Dantas de Lucas, R.; Caputo, F.; Mendes de Souza, K.; Sigwalt, A.R.; Ghisoni, K.; Lock Silveira, P.C.; Remor, A.P.; da Luz Scheffer, D.; Antonacci Guglielmo, L.G.; Latini, A. Increased platelet oxidative metabolism, blood oxidative stress and neopterin levels after ultra-endurance exercise. J. Sports Sci. 2014, 32, 22–30.
- Moser, B.; Schroecksnadel, K.; Hörtnagl, H.; Rieder, J.; Fuchs, D.; Gottardis, M. Influence of Extreme Long Endurance Sports Activity on Neopterin Excretion. Pteridines 2008, 19, 114–119.
- Lindsay, A.; Lewis, J.; Scarrott, C.; Draper, N.; Gieseg, S.P. Changes in acute biochemical markers of inflammatory and structural stress in rugby union. J. Sports Sci. 2015, 33, 882–891.
- Lindsay, A.; Janmale, T.; Draper, N.; Gieseg, S.P. Measurement of changes in urinary neopterin and total neopterin in body builders using SCX HPLC. Pteridines 2014, 25, 53–63.
- Mrakic-Sposta, S.; Gussoni, M.; Vezzoli, A.; Dellanoce, C.; Comassi, M.; Giardini, G.; Bruno, R.M.; Montorsi, M.; Corciu, A.; Greco, F.; et al. Acute Effects of Triathlon Race on Oxidative Stress Biomarkers. Oxid. Med. Cell. Longev. 2020, 2020, 1–14.
- Mrakic-Sposta, S.; Gussoni, M.; Moretti, S.; Pratali, L.; Giardini, G.; Tacchini, P.; Dellanoce, C.; Tonacci, A.; Mastorci, F.; Borghini, A.; et al. Effects of Mountain Ultra-Marathon Running on ROS Production and Oxidative Damage by Micro-Invasive Analytic Techniques. PLoS ONE 2015, 10, e0141780.
- da Luz Scheffer, D.; Latini, A. Exercise-induced immune system response: Anti-inflammatory status on peripheral and central organs. Biochim. Biophys. Acta - Mol. Basis Dis. 2020, 1866, 165823.
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13.
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398.
- Tost, J. DNA methylation: An introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010, 44, 71–81.
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science (80-. ). 1999, 286, 950–952.
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity (Edinb). 2010, 105, 4–13.
- Sawan, C.; Vaissiere, T.; Murr, R.; Herceg, Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutat. Res. 2008, 642, 1–13.
- Loenarz, C.; Schofield, C.J. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem. Biol. 2009, 16, 580–583.
- Guo, Y.; Yu, S.; Zhang, C.; Kong, A.N.T. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 337–349.
- Zhao, F.; Zhang, J.; Chang, N. Epigenetic modification of Nrf2 by sulforaphane increases the antioxidative and anti-inflammatory capacity in a cellular model of Alzheimer’s disease. Eur. J. Pharmacol. 2018, 824, 1–10.
- Liu, Z.-Z.; Zhao, X.-Z.; Zhang, X.-S.; Zhang, M. Promoter DNA demethylation of Keap1 gene in diabetic cardiomyopathy. Int. J. Clin. Exp. Pathol. 2014, 7, 8756–8762.
- Ferrari, L.; Vicenzi, M.; Tarantini, L.; Barretta, F.; Sironi, S.; Baccarelli, A.A.; Guazzi, M.; Bollati, V. Effects of Physical Exercise on Endothelial Function and DNA Methylation. Int. J. Environ. Res. Public Health 2019, 16, 2530.
- Dimauro, I.; Paronetto, M.P.; Caporossi, D. Exercise, redox homeostasis and the epigenetic landscape. Redox Biol. 2020, 35, 101477.
- Wang, D.; Ma, Y.; Yang, X.; Xu, X.; Zhao, Y.; Zhu, Z.; Wang, X.; Deng, H.; Li, C.; Gao, F.; et al. Hypermethylation of the Keap1 gene inactivates its function, promotes Nrf2 nuclear accumulation, and is involved in arsenite-induced human keratinocyte transformation. Free Radic. Biol. Med. 2015, 89, 209–219.
- Wang, R.; An, J.; Ji, F.; Jiao, H.; Sun, H.; Zhou, D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008, 373, 151–154.
- Chen, X.; Zhu, X.; Wei, A.; Chen, F.; Gao, Q.; Lu, K.; Jiang, Q.; Cao, W. Nrf2 epigenetic derepression induced by running exercise protects against osteoporosis. Bone Res. 2021, 9, 15.
- Kang, K.A.; Piao, M.J.; Kim, K.C.; Kang, H.K.; Chang, W.Y.; Park, I.C.; Keum, Y.S.; Surh, Y.J.; Hyun, J.W. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: Involvement of TET-dependent DNA demethylation. Cell Death Dis. 2014, 5, e1183.
- Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Kang, H.K.; Chang, W.Y.; Keum, Y.S.; Hyun, J.W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016, 7, 40594–40620.
- Garcea, R.L.; Alberts, B.M. Comparative Studies of Histone Acetylation in Nucleosomes, Nuclei, and Intact Cells. Biol. Chem. 1980, 255, 11454–11463.
- Fowler, E.; Farb, R.; El-Saidy, S. Distribution of the core histones H2A H2B, H3 and H4 during cell replication. Nucleic Acids Res. 1982, 10, 735–748.
- Yang, X.-J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318.
- Jiang, Y.; Jakovcevski, M.; Bharadwaj, R.; Connor, C.; Schroeder, F.A.; Lin, C.L.; Straubhaar, J.; Martin, G.; Akbarian, S. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J. Neurosci. 2010, 30, 7152–7167.
- Zhong, T.; Ren, F.; Huang, C.S.; Zou, W.Y.; Yang, Y.; Pan, Y.D.; Sun, B.; Wang, E.; Guo, Q.L. Swimming exercise ameliorates neurocognitive impairment induced by neonatal exposure to isoflurane and enhances hippocampal histone acetylation in mice. Neuroscience 2016, 316, 378–388.
- de Meireles, L.C.F.; Bertoldi, K.; Cechinel, L.R.; Schallenberger, B.L.; da Silva, V.K.; Schröder, N.; Siqueira, I.R. Treadmill exercise induces selective changes in hippocampal histone acetylation during the aging process in rats. Neurosci. Lett. 2016, 634, 19–24.
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; Mack, N.J.; Ahmad, N. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 2018, 28, 643–661.
- Xue, F.; Huang, J.; Ding, P.; Zang, H.; Kou, Z.; Li, T.; Fan, J.; Peng, Z.; Yan, W. Nrf2/antioxidant defense pathway is involved in the neuroprotective effects of Sirt1 against focal cerebral ischemia in rats after hyperbaric oxygen preconditioning. Behav. Brain Res. 2016, 309, 1–8.
- Yang, X.; Park, S.-H.; Chang, H.-C.; Shapiro, J.S.; Vassilopoulos, A.; Sawicki, K.T.; Chen, C.; Shang, M.; Burridge, P.W.; Epting, C.L.; et al. Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J. Clin. Invest. 2017, 127, 1505–1516.
- Cao, W.; Hong, Y.; Chen, H.; Wu, F.; Wei, X.; Ying, W. SIRT2 mediates NADH-induced increases in Nrf2, GCL, and glutathione by modulating Akt phosphorylation in PC12 cells. FEBS Lett. 2016, 590, 2241–2255.
- Chen, W.-K.; Tsai, Y.-L.; Shibu, M.A.; Shen, C.-Y.; Chang-Lee, S.N.; Chen, R.-J.; Yao, C.-H.; Ban, B.; Kuo, W.-W.; Huang, C.-Y. Exercise training augments Sirt1-signaling and attenuates cardiac inflammation in D-galactose induced-aging rats. Aging (Albany. NY). 2018, 10, 4166–4174.
- Tunca, U.; Saygin, M.; Ozmen, O.; Aslankoc, R.; Yalcin, A. The impact of moderate-intensity swimming exercise on learning and memory in aged rats: The role of Sirtuin-1. Iran. J. Basic Med. Sci. 2021, 24, 1413–1420.
- Vargas-Ortiz, K.; Pérez-Vázquez, V.; Macías-Cervantes, M.H. Exercise and Sirtuins: A Way to Mitochondrial Health in Skeletal Muscle. Int. J. Mol. Sci. 2019, 20, 2717.
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407.
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283.
- Gao, R.; Wang, L.; Bei, Y.; Wu, X.; Wang, J.; Zhou, Q.; Tao, L.; Das, S.; Li, X.; Xiao, J. Long Noncoding RNA Cardiac Physiological Hypertrophy–Associated Regulator Induces Cardiac Physiological Hypertrophy and Promotes Functional Recovery After Myocardial Ischemia-Reperfusion Injury. Circulation 2021, 144, 303–317.
- Lin, H.; Zhu, Y.; Zheng, C.; Hu, D.; Ma, S.; Chen, L.; Wang, Q.; Chen, Z.; Xie, J.; Yan, Y.; et al. Antihypertrophic Memory After Regression of Exercise-Induced Physiological Myocardial Hypertrophy Is Mediated by the Long Noncoding RNA Mhrt779. Circulation 2021, 143, 2277–2292.
- Qiu, Y.; Zhu, G.; Zeng, C.; Yuan, S.; Qian, Y.; Ye, Z.; Zhao, S.; Li, R. Next-generation sequencing of miRNAs and lncRNAs from rat femur and tibia under mechanical stress. Mol. Med. Rep. 2021, 24, 561.
- Wohlwend, M.; Laurila, P.-P.; Williams, K.; Romani, M.; Lima, T.; Pattawaran, P.; Benegiamo, G.; Salonen, M.; Schneider, B.L.; Lahti, J.; et al. The exercise-induced long noncoding RNA CYTOR promotes fast-twitch myogenesis in aging. Sci. Transl. Med. 2021, 13.
- Zhao, W.; Yin, Y.; Cao, H.; Wang, Y. Exercise Improves Endothelial Function via the lncRNA MALAT1/miR-320a Axis in Obese Children and Adolescents. Cardiol. Res. Pract. 2021, 2021, 1–8.
- Zhang, X.; Wang, Y.; Zhao, Z.; Chen, X.; Li, W.; Li, X. Transcriptome sequencing reveals aerobic exercise training-associated lncRNAs for improving Parkinson’s disease. 3 Biotech 2020, 10, 498.
- Wang, J.; Niu, Y.; Tao, H.; Xue, M.; Wan, C. Knockdown of lncRNA TUG1 inhibits hippocampal neuronal apoptosis and participates in aerobic exercise-alleviated vascular cognitive impairment. Biol. Res. 2020, 53, 53.
- Kazeminasab, F.; Marandi, S.M.; Baharlooie, M.; Safaeinejad, Z.; Nasr-Esfahani, M.H.; Ghaedi, K. Aerobic exercise modulates noncoding RNA network upstream of FNDC5 in the Gastrocnemius muscle of high-fat-diet-induced obese mice. J. Physiol. Biochem. 2021, 77, 589–600.
- He, Y.; Qiang, Y. Mechanism of Autonomic Exercise Improving Cognitive Function of Alzheimer’s Disease by Regulating lncRNA SNHG14. Am. J. Alzheimer’s Dis. Other Dement. 2021, 36, 36.
- Mei, T.; Liu, Y.; Wang, J.; Zhang, Y. miR-340-5p: A potential direct regulator of Nrf2 expression in the post-exercise skeletal muscle of mice. Mol. Med. Rep. 2018, 19, 1340–1348.
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian Folate-Mediated One-Carbon Metabolism. Annu. Rev. Nutr. 2010, 30, 57–81.
- Vineis, P.; Chuang, S.-C.; Vaissière, T.; Cuenin, C.; Ricceri, F.; Collaborators, G.; Johansson, M.; Ueland, P.; Brennan, P.; Herceg, Z. DNA methylation changes associated with cancer risk factors and blood levels of vitamin metabolites in a prospective study. Epigenetics 2011, 6, 195–201.
- Wakefield, L.; Boukouvala, S.; Sim, E. Characterisation of CpG methylation in the upstream control region of mouse Nat2: Evidence for a gene–environment interaction in a polymorphic gene implicated in folate metabolism. Gene 2010, 452, 16–21.
- McKay, J.A.; Xie, L.; Harris, S.; Wong, Y.K.; Ford, D.; Mathers, J.C. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol. Nutr. Food Res. 2011, 55, 1026–1035.
- McKay, J.A.; Wong, Y.K.; Relton, C.L.; Ford, D.; Mathers, J.C. Maternal folate supply and sex influence gene-specific DNA methylation in the fetal gut. Mol. Nutr. Food Res. 2011, 55, 1717–1723.
- Hirsch, S.; Ronco, A.M.; Guerrero-Bosagna, C.; de la Maza, M.P.; Leiva, L.; Barrera, G.; Llanos, M.; Alliende, M.A.; Silva, F.; Bunout, D. Methylation status in healthy subjects with normal and high serum folate concentration. Nutrition 2008, 24, 1103–1109.
- Chen, M.J.; Shimada, T.; Moulton, A.D.; Cline, A.; Humphries, R.K.; Maizel, J.; Nienhuis, A.W. The functional human dihydrofolate reductase gene. J. Biol. Chem. 1984, 259, 3933–3943.
- Holmquist, C.; Larsson, S.; Wolk, A.; de Faire, U. Multivitamin Supplements Are Inversely Associated with Risk of Myocardial Infarction in Men and Women—Stockholm Heart Epidemiology Program (SHEEP). J. Nutr. 2003, 133, 2650–2654.
- Lamprecht, S.A.; Lipkin, M. Chemoprevention of colon cancer by calcium, vitamin D and folate: Molecular mechanisms. Nat. Rev. Cancer 2003, 3, 601–614.
- Smithells, R.W.; Sheppard, S.; Schorah, C.J. Vitamin dificiencies and neural tube defects. Arch. Dis. Child. 1976, 51, 944–950.
- Laurence, K.M.; James, N.; Miller, M.H.; Tennant, G.B.; Campbell, H. Double-blind randomised controlled trial of folate treatment before conception to prevent recurrence of neural-tube defects. BMJ 1981, 282, 1509–1511.
- Yoshida, Y.-I.; Eda, S.; Masada, M. Alterations of tetrahydrobiopterin biosynthesis and pteridine levels in mouse tissues during growth and aging. Brain Dev. 2000, 22, 45–49.
- Bendall, J.K.; Douglas, G.; McNeill, E.; Channon, K.M.; Crabtree, M.J. Tetrahydrobiopterin in Cardiovascular Health and Disease. Antioxid. Redox Signal. 2014, 20, 3040–3077.
- Crabtree, M.J.; Tatham, A.L.; Hale, A.B.; Alp, N.J.; Channon, K.M. Critical Role for Tetrahydrobiopterin Recycling by Dihydrofolate Reductase in Regulation of Endothelial Nitric-oxide Synthase Coupling. J. Biol. Chem. 2009, 284, 28128–28136.
- Bagley, J.R.; Burghardt, K.J.; McManus, R.; Howlett, B.; Costa, P.B.; Coburn, J.W.; Arevalo, J.A.; Malek, M.H.; Galpin, A.J. Epigenetic Responses to Acute Resistance Exercise in Trained vs. Sedentary Men. J. Strength Cond. Res. 2020, 34, 1574–1580.
- Thöny, B. Tetrahydrobiopterin and its functions. In PKU and BH4: Advances in Phenylketonuria and Tetrahydrobiopterin Research; SPS Publications: Heilbronn, Germany, 2006; pp. 503–504.
- Nishida, Y.; Hara, M.; Higaki, Y.; Taguchi, N.; Nakamura, K.; Nanri, H.; Horita, M.; Shimanoe, C.; Yasukata, J.; Miyoshi, N.; et al. Habitual Light-intensity Physical Activity and ASC Methylation in a Middle-aged Population. Int. J. Sports Med. 2019, 40, 670–677.
- Mariathasan, S.; Newton, K.; Monack, D.M.; Vucic, D.; French, D.M.; Lee, W.P.; Roose-Girma, M.; Erickson, S.; Dixit, V.M. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 2004, 430, 213–218.
- Zhang, Y.; Hashimoto, S.; Fujii, C.; Hida, S.; Ito, K.; Matsumura, T.; Sakaizawa, T.; Morikawa, M.; Masuki, S.; Nose, H.; et al. NFκB2 Gene as a Novel Candidate that Epigenetically Responds to Interval Walking Training. Int. J. Sports Med. 2015, 36, 769–775.
