Papillomaviridae is a diverse family of small, non-enveloped DNA viruses, approximately 50–60 nm in diameter that infect all homoeothermic vertebrates including humans. Human papillomavirus (HPV) E6 and E7 oncoproteins are critical for development and maintenance of the malignant phenotype in HPV-induced cancers. These two viral oncoproteins interfere with a plethora of cellular pathways, including the regulation of cell cycle and the control of apoptosis, which are critical in maintaining normal cellular functions. E6 and E7 bind directly with certain components of the Ubiquitin Proteasome System (UPS), enabling them to manipulate a number of important cellular pathways.
- E6
- E7
- HPV
- cervical cancer
- proteasome
- UPS
- ubiquitin ligases
- ubiquitin
1. Introduction
2. The Ubiquitin Proteasome System
3. HPV and the UPS
4. HPV E6 Oncoprotein and the UPS
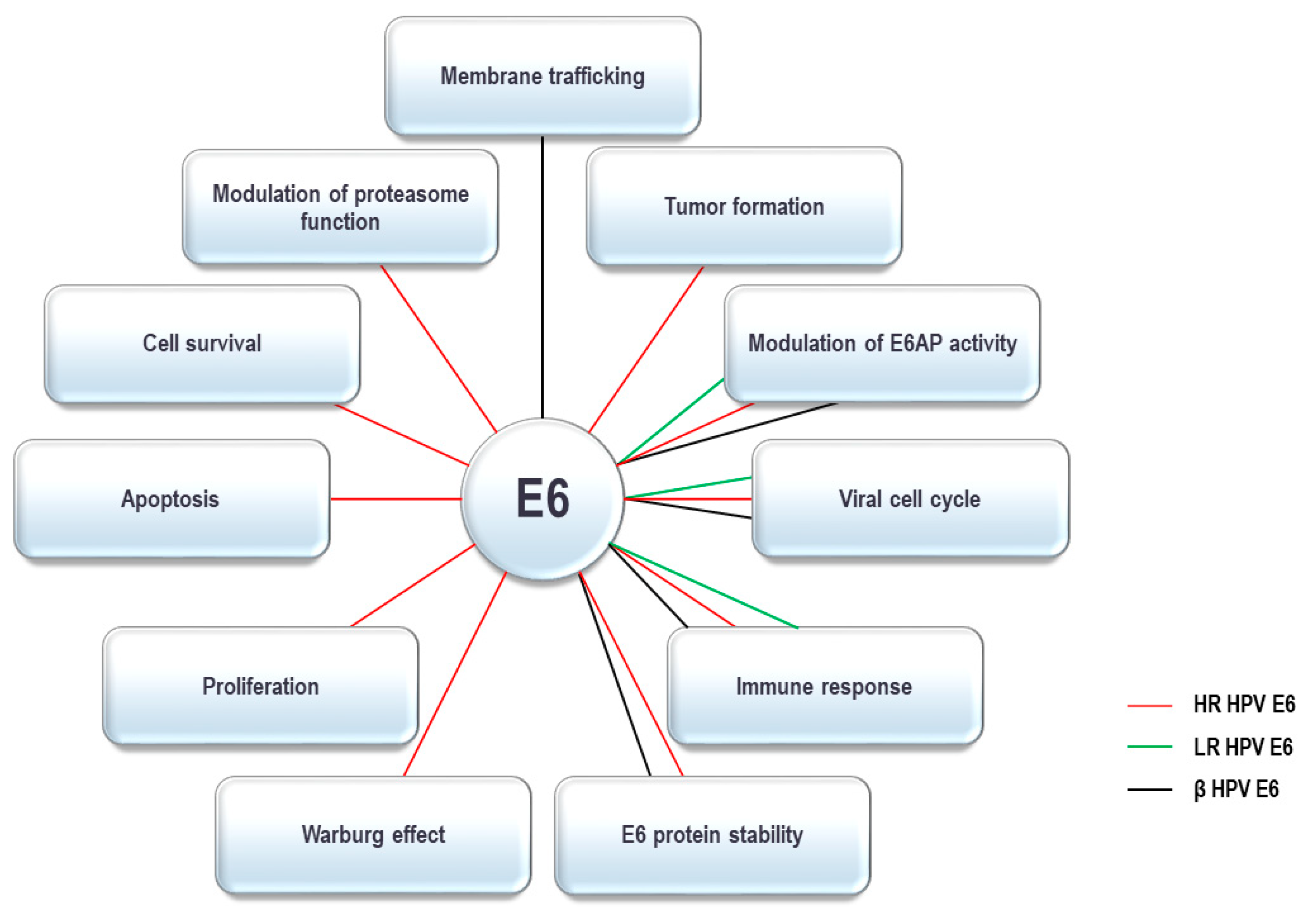
|
Gene ID |
Gene |
Protein |
Function |
α-HPV |
β-HPV |
Method |
Ref. |
|
|---|---|---|---|---|---|---|---|---|
|
LR |
HR |
|||||||
|
580 |
BARD1 |
Breast Cancer 1 Gene (BRCA1)- associated RING domain protein 1 |
Putative tumor suppressor gene mutated cancers. Homologous to BRCA1 RING motif and BRCT domain. BARD1/BRCA1 heterodimer is disrupted by tumorigenic amino acid substitutions in BRCA1. Heterodimer is required for BRCA1 tumor suppression and increases stability of both proteins. |
− |
+ |
? |
Yeast-two hybrid, co-IP |
Yim et al. (2007) |
|
672 |
BRCA1 |
Breast Cancer type 1 susceptibility protein |
Tumor suppressor. The E3 ubiquitin-protein ligase component of BARD1/BRCA1 heterodimer. BRCA1/BARD1 heterodimer coordinates DNA damage repair, ubiquitination and transcriptional regulation to maintain genomic stability. |
? |
+ |
? |
IP, GST-pull down |
Zhang et al. (2005) |
|
4850 |
CNOT4 |
CCR4-NOT transcription complex subunit 4 |
E3 ubiquitin-protein ligase, promoting degradation of target proteins. Involved in JAK/STAT activation |
− |
− |
+ |
IP, MS |
White et al. (2012) |
|
1540 |
CYLD |
CYLD lysine 63 (K63) DUB |
A lysine 63 (K63) deubiquitinase. Tumor suppressor negatively regulating NF-κB pathway. Ubiquitinated and degraded during Hypoxia-induced NF-κB activation to relieve its inhibition of NF-κB signaling cascade |
? |
+ |
? |
An et al. (2008) |
|
|
8925 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
An E3 ubiquitin-protein ligase accepts ubiquitin from an E2 ubiquitin-conjugating enzyme and then transfers the ubiquitin to targeted substrates. Involved in membrane trafficking. |
− |
− |
+ |
IP, proximity ligation in situ assay |
Holloway et al. (2015) |
|
8924 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin- protein ligase 2 |
A putative HECT domain E3 ligase. Involved in protein trafficking and degradation pathways regulating ubiquitin-dependent retention of repair proteins on damaged chromosomes. Recruited to sites of DNA damage in response to ionizing radiation. Promotes DNA damage-induced formation of ‘Lys-63’-linked ubiquitin chains. |
<+ |
+ |
<+ |
IP, MS |
Vos et al. (2009); White et al. (2012) |
|
83737 |
ITCH |
E3 ubiquitin- protein ligase Itchy |
An E3 ubiquitin-protein ligase which is involved in the control of inflammatory signaling pathways. An essential component of an ubiquitin-editing protein complex ensuring the transience of inflammatory signaling pathways by regulating ubiquitin-dependent signaling events. Involved in the cellular antiviral response. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
23295 |
MGRN1 |
Mahogunin ring finger 1 |
Has RING- E3 ubiquitin-protein ligase activity in vitro. Involved in regulation of endosome-to-lysosome trafficking. A negative regulator of hedgehog signaling. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
23024 |
PDZRN3 |
PDZ domain containing ring finger 3 |
A member of the LNX (Ligand of Numb Protein-X) family of RING E3 ubiquitin- protein ligases. Required for vascular morphogenesis and differentiation of adipocytes, osteoblasts and myoblasts. |
− |
+ |
+ |
GST-pull down |
Poirson et al. (2017); Thomas and Banks (2015) |
|
29951 |
PDZRN4 |
PDZ domain containing ring finger 4 |
A member of the LNX family of RING E3 ubiquitin-protein ligases. A suppressor of cell proliferation in human liver cancer cell lines. |
− |
+ |
− |
GPCA, co-IP |
Poirson et al. (2017) |
|
5684 |
PSMA3 |
Proteasome subunit alpha 3 |
Components of the 20S core proteasome complex involved in proteolysis of most cellular proteins. Associated with two 19S regulatory particles, form the 26S proteasome. Involved in ATP- dependent degradation of ubiquitinated proteins. |
+ |
+ |
+ |
IP-MS/MS |
White et al. (2012) |
|
5695 |
PSMB7 |
Proteasome subunit beta 7 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5698 |
PSMB9 |
Proteasome subunit beta 9 |
− |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5700 |
PSMC1 |
Proteasome 26S subunit 4, ATPase 1 |
A component of the 26S proteasome belonging to the heterohexameric ring of AAA proteins (ATPases associated with diverse cellular activities). Unfolds ubiquitinated target proteins. |
? |
+ |
? |
GST-pull down |
Tomaić et al. (2013) |
|
5701 |
PSMC2 |
Proteasome 26S subunit 7, ATPase 2 |
Components of the 26S proteasome. |
+ |
+ |
+ |
IP-MS/MS |
White et al. (2012); Tomaić et al. (2013) |
|
5702 |
PSMC3 |
Proteasome 26S regulatory subunit 6A, ATPase 3 |
+ |
+ |
− |
IP-MS/MS, GST-pull donw |
White et al. (2012); Tomaić et al. (2013) |
|
|
5704 |
PSMC4 |
Proteasome 26S subunit 6B, ATPase 4 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5705 |
PSMC5 |
26S proteasome regulatory subunit 8 |
? |
+ |
? |
GST-pull down |
Tomaić et al. (2013) |
|
|
5707 |
PSMD1 |
26S proteasome non-ATPase regulatory subunit 1 |
+ |
+ |
+ |
IP-MS/MS |
White et al. (2012) |
|
|
5708 |
PSMD2 |
Proteasome 26S subunit 2, non-ATPase 2 |
Component of the 26S proteasome, binds to the intracellular domain of tumor necrosis factor type 1 receptor (TNFR1); the binding domain of TRAP1 and TRAP2 resides outside the death domain of TNFR1. |
+ |
+ |
+ |
IP-MS/MS, GST-pull down |
White et al. (2012); Tomaić et al. (2013) |
|
5710 |
PSMD4 |
26S proteasome non-ATPase regulatory subunit 4 |
A major ubiquitin- accepting proteasome subunit. Involved in maintaining structural integrity of the 19S regulatory particle. Important in direct and indirect recognition of ubiquitinated substrates of 26S proteasome by interacting with polyubiquitinated proteins and directing them to the proteasome for degradation. A critical controlling factor in regulation of protein degradation at the proteasome. |
? |
+ |
? |
IP-MS/MS |
Tomaić et al. (2013) |
|
5709 |
PSMD3 |
Proteasome 26S subunit 3, non-ATPase 3 |
Components of the 26S proteasome. |
+ |
+ |
+ |
IP-MS/MS |
White et al. (2012) |
|
9861 |
PSMD6 |
Proteasome 26S subunit, non-ATPase 6 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5713 |
PSMD7 |
Proteasome 26S subunit, non-ATPase 7 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5714 |
PSMD8 |
Proteasome 26S subunit, non-ATPase 8 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
5719 |
PSMD13 |
26S proteasome non-ATPase regulatory subunit 13 |
+ |
+ |
+ |
IP-MS/MS |
White et al. (2012) |
|
|
10213 |
PSMD14 |
26S proteasome non-ATPase regulatory subunit 14 |
+ |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
|
23198 |
PSME4 |
Proteasome activator subunit 4 |
A proteasome component that specifically recognizes and promotes ATP- and ubiquitin-independent degradation of acetylated core histones during DNA damage response to double-strand breaks. |
− |
+ |
− |
IP-MS/MS |
White et al. (2012) |
|
64320 |
RNF25 |
Ring finger protein 25 |
A RING finger- dependent E3 ubiquitin- protein ligase that mediates ubiquitination and stimulates transcription mediated by NF-κB. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
9810 |
RNF40 |
Ring finger protein 40 |
A component of the RNF20/40 E3 ubiquitin- protein ligase complex; forms a H2B ubiquitin ligase complex in cooperation with the UBE2A or UBE2B. Supports maintenance of tumorigenic features and inflammatory signaling by promoting nuclear NF-κB activity. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
85456 |
TNKS1BP1 |
Tankyrase-1-binding protein |
A subunit of the smaller 1-MDa core of Ccr4-Not complex. Ccr4-Not is an mRNA deadenylase and has a ubiquitin-protein ligase function. |
<+ |
<+ |
+ |
IP-MS/MS |
White et al. (2012) |
|
7188 |
TRAF5 |
TNF receptor associated factor 5 |
Tumor necrosis factor receptor-associated factor (TRAF) protein family. An adapter protein and signal transducer linked to various signaling pathways by association with the receptor cytoplasmic domain and kinases. Involved in cytokine signaling and mediates activation of NF-κB and JNK. It is also involved in apoptosis. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7189 |
TRAF6 |
TNF receptor- associated factor 6 |
An E3 ubiquitin-protein ligase that mediates the synthesis of ‘Lys-63’- linked-polyubiquitin chains conjugated to target proteins and ubiquitination of unanchored poly- ubiquitin chains. Induces activation of NF-κB and JUN. An adaptor protein and signal transducer linking TNFR proteins to different signaling pathways. Plays a role in signal transduction initiated via TNF receptor, IL-1 receptor and IL-17 receptor. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7706 |
TRIM25 |
Tripartite motif containing 25 |
An E3 ubiquitin-protein ligase involved in innate immune defense against viruses. Crucially involved in the interferon response to viral infection. |
+ |
+ |
+ |
co-IP |
Chiang et al. (2018) |
|
10422 |
UBAC1 |
UBA domain containing 1 |
A ubiquitin-protein ligase required for poly-ubiquitination and proteasome-mediated degradation of CDKN1B during G1 phase of the cell cycle. A non-catalytic subunit of the KPC Kip1 ubiquitination- promoting complex. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7337 |
UBE3A |
Ubiquitin- protein ligase E3A |
An E3-HECT domain- containing ubiquitin- protein ligase. It promotes its own degradation in vivo. This imprinted gene is maternally expressed in the brain and biallelically expressed in other tissues. It plays an important role in regulation of the circadian clock and acts as a regulator of synaptic development. |
<+ |
+ |
+ |
Yeast-two hybrid, co-crystal x-ray crystalography, co-IP |
Poirson et al. (2017); Storey et al. (1998); Kao et al. (2000); Brimer et al., 2007; Thomas et al. (2013) |
|
23352 |
UBR4 |
Ubiquitin- protein ligase E3 component n-recognin 4 |
An E3 ubiquitin-protein ligase, a component of the N-end rule pathway. Recognizes and binds to proteins bearing a specific N-terminal leading to ubiquitination and subsequent degradation. Forms meshwork structures involved in membrane morphogenesis and cytoskeletal organization and regulates integrin-mediated signaling. |
− |
− |
+ |
IP |
White et al. (2012); Thomas et al. (2013) |
|
51366 |
UBR5 |
Ubiquitin- protein ligase E3 component n-recognin 5 |
Also known as EDD (E3 identified by Differential Display). A HECT domain-containing E3 ubiquitin-protein ligase component of the N-end rule pathway. Involved in coordinating the balance between cell cycle progression and differentiation. A regulator of DNA damage response, acts as a guard against excessive spreading of ubiquitinated chromatin at damaged chromosomes, as well as tumor suppressor. Frequently overexpressed in breast and ovarian cancer. |
− |
+ |
+ |
co-IP, MS |
Tomaić et al. (2011); White et al. (2012) |
|
9958 |
USP15 |
Ubiquitin specific peptidase 15 |
A deubiquitinating enzyme of the ubiquitin specific protease (USP) family. Plays a critical role in ubiquitin- dependent processes through polyubiquitin chain disassembly and hydrolysis of ubiquitin- substrate bonds. |
− |
+ |
+ |
Yeast-two hybrid, GPCA, IP |
Vos et al. (2009); Poirson et al. (2017); Yaginuma et al. (2018); Chiang et al. (2018) |
|
64854 |
USP46 |
Ubiquitin carboxyl- terminal hydrolase 46 |
A deubiquitinating enzyme that plays a role in behavior by regulating GABA action. Has little intrinsic deubiquitinating activity and requires interaction with regulator of deubiquitinating complexes WDR48 (WD repeat-containing protein 48) for high activity. |
− |
+ |
? |
co-IP |
Kiran et al. (2018) |
|
7428 |
VHL |
Von Hippel-Lindau tumor suppressor |
VHL is a component of the protein complex that includes elongin C, elongin B, and cullin-2. An E3 ubiquitin-protein ligase involved in the ubiquitination and degradation of hypoxia- inducible factor (HIF), a transcription factor crucial to oxygen-related gene expression.. HPV16 E6 promotes hypoxia- induced Warburg effect through blocking the association of HIF-1α and VHL |
? |
+ |
? |
GPCA |
Poirson et al. (2017); Guo et al. (2014) |
|
331 |
XIAP |
X-linked inhibitor of apoptosis |
A multi-functional protein that regulates apoptosis, modulates inflammatory signaling and immunity, cell proliferation, cell invasion and metastasis. An E3 ubiquitin-protein ligase regulating NF-κB signaling and other target. An important regulator of innate immune signaling via regulation of Nod-like receptors (NLRs). |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
+ confirmed to interact; +, < confirmed interactions with lower affinity compared to hR-HPV E6; - interactions not detected; ? interactions not tested. Co-IP: co-immunoprecipitation; GPCA: Gaussia princeps luciferase protein complementation assay, IP-MS/MS: immunoprecipitation-mass spectrometry, IP: immunoprecipitation.
5. HPV E7 Oncoprotein and the UPS
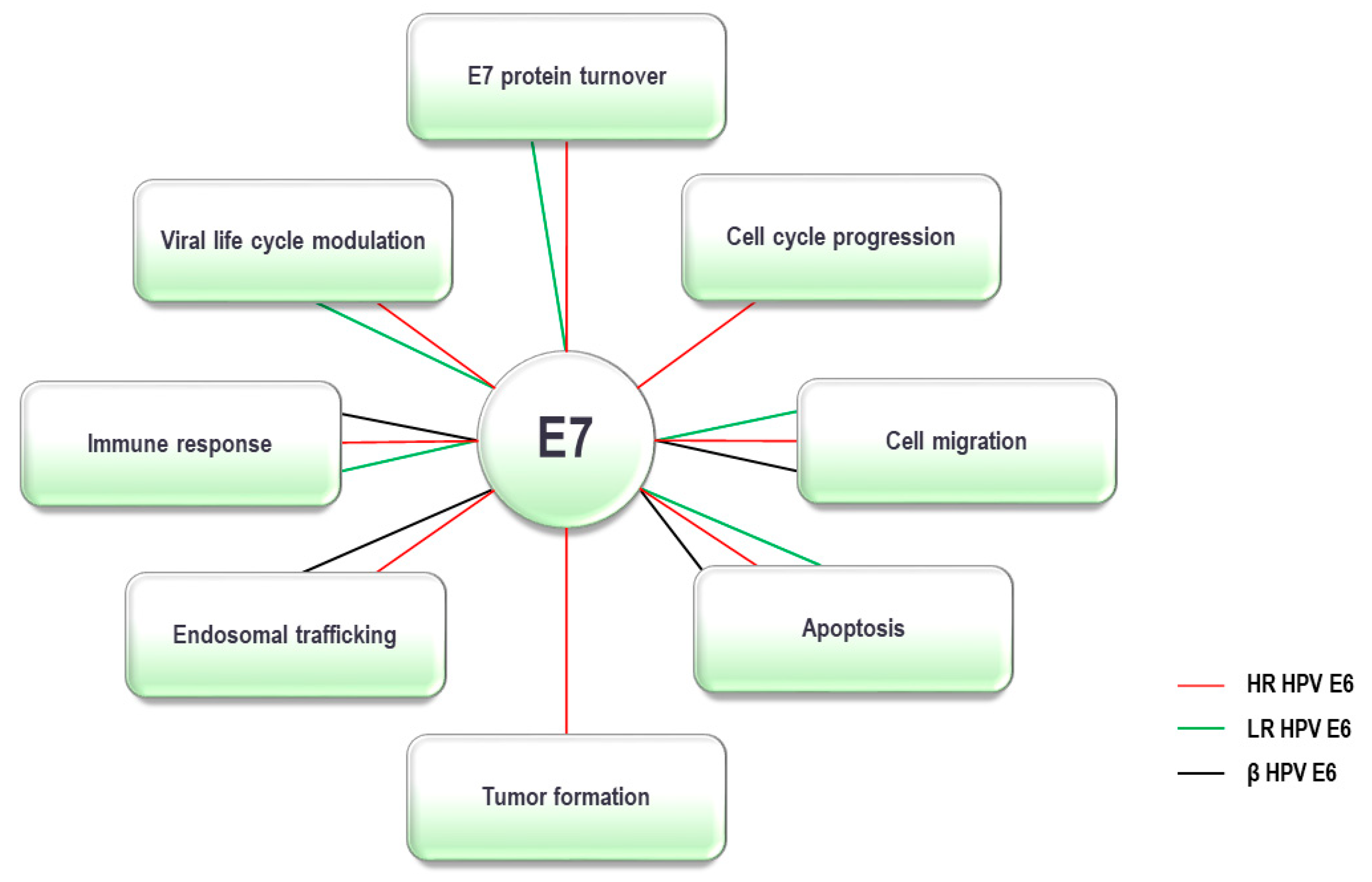
|
Gene ID |
Gene |
Protein |
Function |
α-HPV |
β-HPV |
Method |
Ref. |
|
|---|---|---|---|---|---|---|---|---|
|
LR |
HR |
|||||||
|
8454 |
CUL1 |
Cullin-1 |
A core component of cullin-RING-based SCF E3 ubiquitin- protein ligase complex, which mediates ubiquitination and proteolysis of E7. |
+ |
+ |
? |
IP |
Münger et al. (1989); Boyer et al. (1996); Oh et al. (2004) |
|
8453 |
CUL2 |
Cullin-2 |
A core component of cullin-RING-based ECS E3 ubiquitin- protein ligase complex. Stabilizes APOBEC3A. Required for E7-induced degradation of pRB contributing to cell transformation by dysregulating G1/S cell cycle checkpoints |
− |
+ |
- |
co-IP |
Huh et al. (2007); Narisawa-Saito and Kiyono. (2007); White et al. (2012); Xu et al. (2016); Westrich et al. (2018) |
|
8452 |
CUL3 |
Cullin-3 |
A core component of cullin-RING-based BCR E3 ubiquitin- protein ligase complexes, which mediate the ubiquitination and proteasomal degradation of target proteins. |
+ |
+ |
? |
co-IP |
White et al. (2012); Poirson et al. (2017) |
|
90379 |
DCAF15 |
DDB1 and CUL4 associated factor 15 |
May be involved in ubiquitination and degradation through a DBB1-CUL4 E3 protein-ubiquitin ligase. |
− |
+ |
− |
GPCA, co-IP |
Poirson et al. (2017) |
|
253980 |
KCTD13 |
Potassium channel tetramer- ization domain 13 |
A substrate-specific adapter of a BCR E3 ubiquitin-protein ligase complex required for synaptic transmission. |
+ |
+ |
+ |
GPCA, co-IP |
Chen et al. (2009); Poirson et al. (2017) |
|
112939 |
NACC1 |
Nucleus accumbens- associated protein 1 |
A transcriptional repressor and transcriptional corepressor in neuronal cells through recruitment of HDAC3 and HDAC4 Required to recruit the proteasome from the nucleus to the cytoplasm and dendritic spines. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
9148 |
NEURL1 |
Neuralized E3 ubiquitin- protein ligase 1 |
An E3 ubiquitin- protein ligase that activates in vitro ubiquitination of JAG1, inhibiting malignant cell transformation of medulloblastoma cells through the Notch pathway. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
5700 |
PSMC1 |
26S proteasome regulatory subunit 4 |
A component of the 26S proteasome. |
− |
+ |
? |
IP |
Berezutskaya and Bagchi (1997); Ben-Saadon et al. (2004) |
|
84282 |
RNF135 |
ring finger protein 135 |
An E2-dependent E3 ubiquitin-protein ligase, involved in innate immune defense against viruses. |
+, < |
+ |
− |
GPCA, co-IP |
Poirson et al. (2017) |
|
57630 |
SH3RF1 |
SH3 domain containing ring finger 1 |
Has an E3 ubiquitin-protein ligase activity. In the absence of an external substrate, it can catalyze self-ubiquitination. |
+, < |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
92799 |
SHKBP1 |
SH3KBP1- binding protein 1 |
SHKBP1 inhibits CBL-SH3KBP1 complex mediated downregulation of EGFR signaling by sequestration of SH3KBP1. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
7126 |
TNFAIP1 |
TNF alpha induced protein 1 |
A substrate-specific adapter of a BCR E3 ubiquitin-protein ligase complex that mediates the ubiquitination and proteasomal degradation of RhoA, thereby regulating the actin cytoskeleton and cell migration. HPV-16 E7 can modulate the responses of its natural host cell to the closely related cytokines TNF-α. |
+ |
+ |
+ |
GPCA, co-IP |
Basile et al. (2001); Poirson et al. (2017) |
|
7186 |
TRAF2 |
TNF receptor associated factor 2 |
Regulates the activation of NF-κB and JNK and plays a central role in the regulation of cell survival and apoptosis. An essential constituent of several E3 ubiquitin-protein ligases. HPV16 E6/E7 switch cells from apoptotic to proliferative fates under TWEAK/Fn14 interaction, possibly by favoring Ras and TRAF2 activation and modulating TNF receptor expression. |
− |
+ |
+ |
GPCA, co-IP |
Cheng et al. (2015); Poirson et al. (2017) |
|
7187 |
TRAF3 |
TNF receptor associated factor 3 |
Regulates pathways leading to activation of NF-κB and MAP kinases, and plays a central role in the regulation of B-cell survival. An essential constituent of several E3 ubiquitin-protein ligase complexes. Overexpression of TRAF3 enhances p53 and pRb expression |
? |
+ |
? |
GPCA |
Poirson et al. (2017); Zhang et al. (2018) |
|
9618 |
TRAF4 |
TNF receptor associated factor 4 |
An adaptor protein and signal transducer linking members of the TNFR family to different signaling pathways. Plays a role in the activation of NF-κB and JNK, and in the regulation of cell survival and apoptosis. May interact selectively and non-covalently with E3 ubiquitin- protein ligase enzymes. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7188 |
TRAF5 |
TNF receptor associated factor 5 |
An adaptor protein and signal transducer linking members of the TNFR family to different signaling pathways. May interact selectively and non-covalently with E3 ubiquitin-protein ligase enzymes. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
10346 |
TRIM22 |
tripartite motif containing 22 |
An interferon- induced antiviral protein involved in innate immunity. May have E3 ubiquitin-protein ligase activity. Activated by integration of E6/E7 genes. |
? |
+ |
? |
GPCA |
Pett et al. (2006); Poirson et al. (2017) |
|
22954 |
TRIM32 |
tripartite motif containing 32 |
An E3 ubiquitin-protein ligase. It ubiquitinates DTNBP1 and promotes its degradation. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
493829 |
TRIM72 |
tripartite motif containing 72 |
A muscle-specific protein that plays a central role in cell membrane repair by nucleating the assembly of the repair machinery at injury sites. May be involved in proteasome- mediated, ubiquitin-dependent protein catabolic processes. |
− |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
114088 |
TRIM9 |
tripartite motif containing 9 |
An E3 ubiquitin- protein ligase, which self-ubiquitinates in cooperation with E2 enzyme UBE2D2/UBC4. Serves as a targeting signal for proteasomal degradation. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7314 |
UBB |
ubiquitin B |
Targets cellular proteins for degradation by the 26S proteasome. Involved in the maintenance of chromatin structure, the regulation of gene expression, and the stress response. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7319 |
UBE2A |
Ubiquitin- conjugating enzyme E2A |
Accepts ubiquitin from the E1 complex. In association with the E3 enzyme, UBE2A plays a role in transcription regulation by catalyzing the ubiquitination of histone H2B. |
+ |
+ |
? |
GPCA |
Poirson et al. (2017) |
|
7332 |
UBE2L3 |
Ubiquitin- conjugating enzyme E2L3 |
Specifically acts with HECT-type and RBR family E3 ubiquitin- protein ligases. Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins including E7. |
? |
+ |
? |
IP |
Reinstein et al. (2000) |
|
23352 |
UBR4 |
Ubiquitin- protein ligase E3 component n-recognin 4 |
Also known as p600. An E3 ubiquitin- protein ligase that recognizes proteins with specific destabilized N-terminal residues, leading to their ubiquitination and degradation. May mediate some pRB- independent transforming activities of HPV-16 E7, but is not sufficient for cellular transformation as interactions were also found with low-risk HPV E7 oncoproteins. It is speculated that UBR4 could potentially play a role in viral replication. |
+ |
+ |
+ |
IP |
Huh et al. (2005); De Masi et al. (2005); White et al. (2012) |
|
8237 |
USP11 |
Ubiquitin carboxyl- terminal hydrolase 11 |
Inhibits degradation of target proteins by the proteasome. Plays a role in the regulation of pathways leading to NF-κ-B activation. Augments HPV-16E7 activity in modulating downstream target genes, such as pRb, Bcl-2, and Cdc-2, suggesting that this interaction may contribute to cell transformation by HPV-16E7. |
? |
+ |
? |
IP |
Lin et al. (2008); Poirson et al. (2017) |
|
83844 |
USP26 |
Ubiquitin carboxyl- terminal hydrolase 26 |
Involved in the ubiquitin-dependent proteolytic pathway in conjunction with the 26S proteasome. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
57663 |
USP29 |
Ubiquitin carboxyl- terminal hydrolase 29 |
A thiol-dependent hydrolyser of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin. |
− |
+ |
− |
GPCA, co-IP |
Poirson et al. (2017) |
|
23032 |
USP33 |
Ubiquitin carboxyl- terminal hydrolase 33 |
A deubiquitinating enzyme involved in centrosome duplication, cell migration and beta-2 adrenergic receptor/ADRB2 recycling. |
+ |
+ |
+ |
GPCA, co-IP |
Poirson et al. (2017) |
|
90850 |
ZNF598 |
Zinc finger protein 598 |
An E3 ubiquitin- protein ligase required for terminal stalling of ribosomes during translation of poly(A) sequences by mediating ubiquitination of 40S ribosomal protein. |
? |
+ |
? |
GPCA |
Poirson et al. (2017) |
+ confirmed to interact; +, < confirmed interactions with lower affinity compared to HR-HPV E7; - interactions not detected; ? interactions not tested. Co-IP: co-immunoprecipitation; GPCA: Gaussia princeps luciferase protein complementation assay, IP-MS/MS: immunoprecipitation-mass spectrometry, IP: immunoprecipitation.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens9020133
References
- International Agency for Cancer Research. IARC Monographs 100B—Human Papillomaviruses 2012; International Agency for Cancer Research, IARC Press: Lyon, France, 2005.
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy: Classification and Nomenclature of Viruses. Virus Taxon. 2005.
- López-Bueno, A.; Mavian, C.; Labella, A.M.; Castro, D.; Borrego, J.J.; Alcami, A.; Alejo, A. Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream. J. Virol. 2016, 90, 8768–8779.
- de Villiers, E.-M.; Burk, R.D.; Bernard, H.-U.; Chen, Z.; van Doorslaer, K.; zur Hausen, H. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79.
- Doorbar, J. The papillomavirus life cycle. J. Clin. Virol. 2005, 32 (Suppl. S1), S7–S15.
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560.
- Morgan, E.L.; Wasson, C.W.; Hanson, L.; Kealy, D.; Pentland, I.; McGuire, V.; Roberts, S. STAT3 activation by E6 is essential for the differentiation-dependent HPV18 life cycle. PLoS Pathog. 2018, 14.
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1997, 425–479.
- Ciechanover, A. Intracellular protein degradation: From a Vague Idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel Lecture). Angew. Chem. Int. Ed. 2005, 44, 5944–5967.
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B 2009, 85, 12–36.
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712.
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124.
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513.
- Poirson, J.; Biquand, E.; Straub, M.-L.; Cassonnet, P.; Nominé, Y.; Jones, L.; Demeret, C. Mapping the interactome of HPV E6 and E7 oncoproteins with the ubiquitin-proteasome system. FEBS J. 2017, 284, 3171–3201.
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984.
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487.
- Li, W.; Ye, Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell. Mol. Life Sci. 2008, 65, 2397–2406.
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409.
- Wing, S.S. Deubiquitinating enzymes-the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 2003, 35, 590–605.
- Tomaić, V.; Banks, L. Angelman syndrome-associated ubiquitin ligase UBE3A/E6AP mutants interfere with the proteolytic activity of the proteasome. Cell Death Dis. 2015, 6, e1625.
- Lou, Z.; Wang, S. E3 ubiquitin ligases and human papillomavirus-induced carcinogenesis. J. Int. Med. Res. 2014, 42, 247–260.
- Vos, R.M.; Altreuter, J.; White, E.A.; Howley, P.M. The Ubiquitin-Specific Peptidase USP15 Regulates Human Papillomavirus Type 16 E6 Protein Stability. J. Virol. 2009, 83, 8885–8892.
- Tomaić, V.; Pim, D.; Banks, L. The stability of the human papillomavirus E6 oncoprotein is E6AP dependent. Virology 2009, 393, 7–10.
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136.
- Helt, A.-M.; Funk, J.O.; Galloway, D.A. Inactivation of both the Retinoblastoma Tumor Suppressor and p21 by the Human Papillomavirus Type 16 E7 Oncoprotein Is Necessary To Inhibit Cell Cycle Arrest in Human Epithelial Cells. J. Virol. 2002, 76, 10559–10568.
- Münger, K.; Werness, B.A.; Dyson, N.; Phelps, W.C.; Harlow, E.; Howley, P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989, 8, 4099–4105.
- Werness, B.A.; Levine, A.J.; Howley, P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990, 248, 76–79.
- Pietsch, E.C.; Murphy, M.E. Low risk HPV-E6 traps p53 in the cytoplasm and induces p53-dependent apoptosis. Cancer Biol. Ther. 2008, 7, 1916–1918.
- Oh, S.T.; Longworth, M.S.; Laimins, L.A. Roles of the E6 and E7 Proteins in the Life Cycle of Low-Risk Human Papillomavirus Type 11. J. Virol. 2004, 78, 2620–2626.
- Crook, T.; Tidy, J.A.; Vousden, K.H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 1991, 67, 547–556.
- Lechner, M.S.; Laimins, L.A. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J. Virol. 1994, 68, 4262–4273.
- White, E.A.; Walther, J.; Javanbakht, H.; Howley, P.M. Genus beta human papillomavirus E6 proteins vary in their effects on the transactivation of p53 target genes. J. Virol. 2014, 88, 8201–8212.
- White, E.A.; Sowa, M.E.; Tan, M.J.A.; Jeudy, S.; Hayes, S.D.; Santha, S.; Münger, K.; Harper, J.W.; Howley, P.M. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. USA 2012, 109, E260–E267.
- Huh, K.; Zhou, X.; Hayakawa, H.; Cho, J.-Y.; Libermann, T.A.; Jin, J.; Harper, J.W.; Munger, K. Human Papillomavirus Type 16 E7 Oncoprotein Associates with the Cullin 2 Ubiquitin Ligase Complex, Which Contributes to Degradation of the Retinoblastoma Tumor Suppressor. J. Virol. 2007, 81, 9737–9747.
- Szalmás, A.; Tomaić, V.; Basukala, O.; Massimi, P.; Mittal, S.; Kónya, J.; Banks, L. The PTPN14 Tumor Suppressor Is a Degradation Target of Human Papillomavirus E7. J. Virol. 2017, 91.
- Holloway, A.; Simmonds, M.; Azad, A.; Fox, J.L.; Storey, A. Resistance to UV-induced apoptosis by β-HPV5 E6 involves targeting of activated BAK for proteolysis by recruitment of the HERC1 ubiquitin ligase. Int. J. Cancer 2015, 136, 2831–2843.
- Bennett Saidu, N.E.; Filić, V.; Thomas, M.; Sarabia-Vega, V.; Ðukić, A.; Miljković, F.; Banks, L.; Tomaić, V. PDZ Domain-Containing Protein NHERF-2 is a Novel Target of Human Papillomavirus type 16 (HPV-16) and HPV-18. J. Virol. 2019.
- Huh, K.-W.; DeMasi, J.; Ogawa, H.; Nakatani, Y.; Howley, P.M.; Münger, K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 2005, 102, 11492–11497.
- DeMasi, J.; Huh, K.-W.; Nakatani, Y.; Münger, K.; Howley, P.M. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc. Natl. Acad. Sci. USA 2005, 102, 11486–11491.
- Tomaić, V. Functional Roles of E6 and E7 Oncoproteins in HPV-Induced Malignancies at Diverse Anatomical Sites. Cancers 2016, 8, 95.
- Grm, H.S.; Banks, L. Degradation of hDlg and MAGIs by human papillomavirus E6 is E6-AP-independent. J. Gen. Virol. 2004, 85, 2815–2819.
- Cole, S.T.; Danos, O. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. Phylogeny of papillomaviruses and repeated structure of the E6 and E7 gene products. J. Mol. Biol. 1987, 193, 599–608.
- Barbosa, M.S.; Wettstein, F.O. Transcription of the cottontail rabbit papillomavirus early region and identification of two E6 polypeptides in COS-7 cells. J. Virol. 1987, 61, 2938–2942.
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Pol, S.V.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545.
- Kanda, T.; Watanabe, S.; Zanma, S.; Sato, H.; Furuno, A.; Yoshiike, K. Human papillomavirus type 16 E6 proteins with glycine substitution for cysteine in the metal-binding motif. Virology 1991, 185, 536–543.
- Sherman, L.; Schlegel, R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J. Virol. 1996, 70, 3269–3279.
- Nominé, Y.; Masson, M.; Charbonnier, S.; Zanier, K.; Ristriani, T.; Deryckère, F.; Sibler, A.P.; Desplancq, D.; Atkinson, R.A.; Weiss, E.; et al. Structural and functional analysis of E6 oncoprotein: Insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell. 2006, 21, 665–678.
- Zanier, K.; Ruhlmann, C.; Melin, F.; Masson, M.; Ould M’hamed Ould Sidi, A.; Bernard, X.; Fischer, B.; Brino, L.; Ristriani, T.; Rybin, V.; et al. E6 proteins from diverse papillomaviruses self-associate both in vitro and in vivo. J Mol Biol 2010, 396, 90–104.
- Zanier, K.; Charbonnier, S.; Sidi, A.O.M.O.; McEwen, A.G.; Ferrario, M.G.; Poussin-Courmontagne, P.; Cura, V.; Brimer, N.; Babah, K.O.; Ansari, T.; et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science 2013, 339, 694–698.
- Pim, D.; Banks, L. Interaction of viral oncoproteins with cellular target molecules: Infection with high-risk vs low-risk human papillomaviruses. APMIS 2010, 118, 471–493.
- Zhang, Y.; Dasgupta, J.; Ma, R.Z.; Banks, L.; Thomas, M.; Chen, X.S. Structures of a human papillomavirus (HPV) E6 polypeptide bound to MAGUK proteins: Mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J. Virol. 2007, 81, 3618–3626.
- Thomas, M.; Narayan, N.; Pim, D.; Tomaić, V.; Massimi, P.; Nagasaka, K.; Banks, L. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 2008, 27, 7018–7030.
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 1993, 13, 4918–4927.
- Elston, R.C.; Napthine, S.; Doorbar, J. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP and E6BP. J. Gen. Virol. 1998, 79 Pt 2, 371–374.
- White, E.A.; Kramer, R.E.; Tan, M.J.A.; Hayes, S.D.; Harper, J.W.; Howley, P.M. Comprehensive Analysis of Host Cellular Interactions with Human Papillomavirus E6 Proteins Identifies New E6 Binding Partners and Reflects Viral Diversity. J. Virol. 2012, 86, 13174–13186.
- Brimer, N.; Lyons, C.; Wallberg, A.E.; Vande Pol, S.B. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene 2012, 31, 4639–4646.
- Tan, M.J.A.; White, E.A.; Sowa, M.E.; Harper, J.W.; Aster, J.C.; Howley, P.M. Cutaneous β-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1473–E1480.
- Meyers, J.M.; Spangle, J.M.; Munger, K. The human papillomavirus type 8 E6 protein interferes with NOTCH activation during keratinocyte differentiation. J. Virol. 2013, 87, 4762–4767.
- Clemens, K.E.; Brent, R.; Gyuris, J.; Munger, K. Dimerization of the human papillomavirus E7 oncoprotein in vivo. Virology 1995, 214, 289–293.
- Münger, K.; Grace, M.; Yee, C.; Jones, D.L.; Mavromatis, K.O.; Mukherjee, R. The carboxyl-terminal zinc-binding domain of the human papillomavirus E7 protein can be functionally replaced by the homologous sequences of the E6 protein. Virus Res. 2002, 52, 109–118.
- Phelps, W.C.; Yee, C.L.; Münger, K.; Howley, P.M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 1988, 53, 539–547.
- Phelps, W.C.; Munger, K.; Yee, C.L.; Barnes, J.A.; Howley, P.M. Structure-Function Analysis of the Human Papillomavirus Type 16 E7 Oncoprotein. J. Virol. 1992, 66, 2418–2427.
- McLaughlin, M.; Münger, K. The Human Papillomavirus E7 Oncoprotein. Virology 2010, 384, 335–344.
- Roman, A.; Munger, K. The papillomavirus E7 proteins. Virology 2013, 445, 138–168.
- Smotkin, D.; Wettstein, F.O. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 1986, 83, 4680–4684.
- Morris, E.J.; Dyson, N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001, 82, 1–54.
