Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Human metapneumovirus (HMPV) is an important human pathogen that, along with respiratory syncytial virus (RSV), is a major cause of respiratory tract infections in young infants. Recombinant chimeric viruses expressing antigens of other viruses can be generated by reverse genetics and used for simultaneous immunization against more than one pathogen. This approach can result in the development of promising vaccine candidates against HMPV, and several studies have indeed validated viral vectors expressing HMPV antigens.
- human metapneumovirus
- respiratory syncytial virus
- chimeric vaccines
- recombinant vaccines
- viral vectors
- bivalent vaccines
1. Introduction
Acute respiratory tract infections (RTIs) are the 5th leading global cause of mortality among all age groups and the 3rd leading mortality cause in children younger than 5 years old [1]. An important number of RTIs is caused by viruses belonging to the Pneumoviridae family, namely, respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) that are responsible for approximately 31% and 5.5% of RTIs in children, respectively [2].
HMPV is a single-stranded, negative-sense RNA virus belonging to the family of Pneumoviridae, genus Metapneumovirus. The genus has two species: human and avian metapneumovirus (AMPV). HMPVs are divided into two major genetic subtypes: A and B that are further divided into sublineages A1, A2a, A2b, B1, and B2 [3]. Lineages A and B share 80% and 90% of their nucleotide and amino acid sequence identity, correspondingly [4]. The major antigen of HMPV, the F protein, is highly conserved and a high level of cross-neutralization and cross-protection is observed between the two lineages [5,6]. The clinical signs of HMPV disease do not vary significantly between the different genetic lineages of HMPV [7]. AMPV, previously known as turkey rhinotracheitis virus, is an important poultry pathogen, divided into subgroups A, B, C, and D [8,9,10]. The subgroup C (AMPV_C) shares 80% of the amino acid identity of N, P, M, F, M2-1, and M2-2 proteins with HMPV [11], making it the closest related virus to HMPV.
HMPV is a relatively newly-discovered pathogen, described for the first time by the researchers from the Netherlands in 2001 [11]. Its late identification can be explained by the similarity of symptoms caused by HMPV and RSV, as well as the difficulty to observe HMPV growth in vitro. Both viruses cause pneumonia and bronchiolitis, with variable severity of illness according to age [12]. Although HMPV disease is generally less severe compared to RSV, the incidence of infections caused by HMPV is similar to that of influenza and parainfluenza, and it causes a significant number of hospitalizations every year [13]. Infections with either virus are associated with the development of asthma and its exacerbations, yet the causal relationship has not been established [2,14,15]. Although both pneumoviruses can infect any age group, they cause the greatest disease severity in young infants. HMPV infects mainly children between 6 and 12 months of age, whereas RSV infects infants earlier, within the first 2–3 months after the birth [2,16,17,18,19]. Virtually every child is infected by RSV by the age of 2 [20] and by HMPV by the age of 5 [21], and the incidence of reinfections with both viruses increases significantly after the age of 50 causing a lot of RTIs in elderly [19,22]. Another population at risk of HMPV infections is comprised of immunosuppressed patients [23].
1.1. Need for a Vaccine
Despite the high frequency of pneumoviral infections and over 50 years of research in this field, no licensed vaccine against HMPV or RSV is currently available. Among the numerous vaccine candidates against RSV that have been developed, only a few have advanced to clinical trials and most of them failed as a result of insufficient immunogenicity or underattenuation [24]. The only anti-HMPV vaccine advanced to clinical trials until now—a live-attenuated recombinant rHMPV-PA virus, in which HMPV-P protein was exchanged for its counterpart from AMPV_C—proved to be insufficiently immunogenic and overattenuated in seronegative children, thus leaving no advanced candidates for an anti-HMPV vaccine [25].
This lack of effective vaccine candidates against HMPV can be explained by the recent discovery of the virus, but also by the lack of a successful vaccine against closely related RSV that could serve as a base for vaccine design. One of the reasons for the slow progress in this field is the clinical failure of formalin-inactivated RSV-vaccine (FI-RSV) that occurred in the 1960s. The administration of FI-RSV led to the development of an exaggerated immune response to wild type (wt) RSV infection with enhanced pulmonary disease (EPD) and two vaccinated children died [26]. EPD has been further documented in various animal models, indicating that it is not only a human phenomenon [27,28,29,30]. A similar effect was observed for a formalin or heat-inactivated HMPV vaccine, which induced the symptoms of EPD in rodents [31,32]. Another reason for slow vaccine development is the transient immunity provided by natural infection with RSV and HMPV. As documented in RSV-seropositive adults, levels of RSV-neutralizing antibodies correlate with the resistance to subsequent RSV infection, but the protection they confer is incomplete and short-lasting, which results in frequent reinfections [20,33]. The immune response to primary HMPV infection was described to be weak and aberrant in BALB/c mice, with excessive Th2-cytokines production at later stages of infection, which correlated with airways hyperresponsiveness and the development of asthma [34]. Pneumoviruses have also developed many mechanisms to evade immune responses. The RSV nonstructural proteins NS1 and NS2 suppress the IFN-induced antiviral response in infected cells and removal of these proteins resulted in the induction of high levels of IFN alpha and beta in vitro [35,36,37]. HMPV shares with RSV the same ability to decrease IFN response, despite the lack of NS1 and NS2 proteins. It has been documented that HMPV-G protein has the ability to inhibit the IFN type 1 response in vitro and in vivo [38,39] and that its SH and M2-2 proteins can also modulate the host’s immune responses [40,41,42]. Both HMPV and RSV can persist in the lungs of an infected animal host, despite the presence of neutralizing antibodies [34,43]. Such persistence has been documented by RT-PCR detection of viral RNA in the guinea pig and mouse models for RSV [44,45,46] and in mice for HMPV [43]. Another hurdle to effective immunization is the presence of maternal antibodies in the bloodstream of vaccinated infants that can impair the immunogenic effect of the vaccine [47], yet the role of maternal antibodies in RSV infection is not fully understood [48].
1.2. Vaccine Development
The main objectives in pneumovirus vaccine development are to avoid EPD and obtain sufficient immunogenicity without causing disease. Viral protein antigens can be used as subunit vaccines to induce an appropriate immune response. These proteins can be delivered as nanoparticles, virus-like particles (VLPs), or they can be coupled with adjuvants [49]. Another interesting strategy is to immunize with viral nucleic acid coding for viral antigens, as it has been documented for RSV and HMPV vaccine candidates [50,51,52]. An mRNA-based vaccine coding for the F proteins of HMPV and human parainfluenza type 3 virus (PIV3) has been recently advanced to phase 1 clinical trial [53].
Among the three surface proteins of HMPV (F, G, and SH), the F protein constitutes the major HMPV antigen [6,54,55]. Although immunogenic, its G protein does not induce a potent protective immune response [54,56] and is not indispensable for viral replication in vivo [57]. HMPV-SH protein was shown not to confer any significant protection [54], the same being observed for RSV [58]. The F protein is also the major antigen of RSV, more immunogenic than the G protein [59], although the latter is also able to induce a protective immune response [60,61] and has been frequently tested as a subunit vaccine [62,63,64]. RSV-F protein stabilized in its pre-fusion form by introducing disulfide bond (DS) and cavity-filling (Cav1) mutations (DS-Cav1) is currently being tested as a subunit vaccine in phase I clinical trials (ClinicalTrials.gov Identifier: NCT03049488). The pre-fusion RSV-F was shown to be more immunogenic than its postfusion conformation, as a result of an exposition of a unique antigenic site Ø that is recognized by very potent RSV-neutralizing antibodies [65,66,67]. Trials with the HMPV-F subunit vaccine showed that the vaccine was immunogenic, but not protective in a rodent model [68]. To increase its immunogenic potential, HMPV-F protein has been stabilized in its pre-fusion state by analogous strategies as for RSV-F, but its immunogenicity was not enhanced [69,70]. This difference between pre-fusion RSV-F and HMPV-F can be explained by additional glycosylation at the apex of the pre-fusion form of HMPV-F. HMPV-F and RSV-F share some antigenic sites, and antibodies able to cross-neutralize the two pneumoviruses were identified [71,72,73,74,75]. Grafting of a major protective epitope from RSV-F into HMPV-F protein resulted in the elaboration of a chimeric protein carrying epitopes of both pneumoviruses, an interesting candidate for vaccine design [76,77].
Several promising nanoparticle vaccines against RSV have been elaborated [78,79], among which is ResVax, a nanoparticle RSV-F-based vaccine developed by Novavax [80]. ResVax has been recently tested in phase 3 clinical trials in maternal immunization model of lower RTIs prevention in infants (NCT02624947), where it did not meet its clinical endpoint [81]. VLPs are structures composed of the proteins of viral capsid; they do not contain the viral genome and are replication-incompetent. Coexpression of HMPV-F, -G, and -M proteins leads to the formation of VLPs in vitro [82], yet the expression of only F and M proteins has been shown sufficient for VLPs assembly [83]. Several potential VLPs vaccines against HMPV [82,83,84,85], and many more against RSV [78,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105], were developed, but none of them has been advanced to clinical trials. A promising vaccine candidate—a VLP composed of pre-fusion, postfusion RSV-F, or both together, assembled with HMPV-M protein—conferred full protection against RSV infection in immunized mice [96].
On the other hand, immunization with an attenuated replication-competent virus is a very promising approach. In this case, it is possible to obtain a vaccine that presents all viral epitopes and is able to induce both humoral and cellular immune responses [106]. The main disadvantage of live attenuated vaccines is the risk of reversion of the attenuated profile, restoration of infectivity and subsequent development of the disease [107]. Live attenuated viruses can be divided into two groups: non-recombinant and recombinant or chimeric viruses. Non-recombinant viruses are rendered less infectious due to genetic modifications that appear naturally during serial passages in vitro in conditions of environmental stress. Several attenuated strains of RSV and HMPV have been obtained as a result of serial cold passages or chemical mutagenesis, which confer a temperature-sensitive (ts) phenotype to a virus [108,109,110,111,112]. Apart from the mentioned selection strategy, recombinant attenuated viruses can be generated by reverse genetics, a technique that makes it possible to generate functional particles of a modified virus based on its genetic material [113,114]. This approach allows not only to introduce attenuating mutations directly into the viral genome [57,115], but also to express exogenous antigens in the backbone of the virus, leading to the development of polyvalent, chimeric vaccines. The insertion of a foreign gene can have a potentially attenuating effect on its own, for example, by rearranging the order of genes in terms of 3’-5’ expression gradient, as it is observed in paramyxoviruses. Gene order can also be changed in a more invasive way by moving virulence genes from the high-expression position to the low-expression locus. Alternatively, the replication efficiency can be decreased by deletions or silencing of nonessential viral proteins that play an accessory function in the life cycle of the virus. It is also possible to swap some of the genes between strains of different host preferences, thus introducing new host range restrictions. HMPV virus with the P gene exchanged for its AMPV_C counterpart (rHMPV-PA) was more attenuated than wt HMPV, and yet protective against HMPV infection in African green monkeys (AGMs) [116].
1.3. Target Populations
The ultimate goal of HMPV vaccination is the prevention of lower RTIs in populations at risk. Similarly to RSV, the target populations for HMPV vaccination are young children, elderly people, and pregnant women [117]. Early vaccination of infants and young children can potentially prevent HMPV-infections and the transmission of the virus. Replication-competent vaccines, namely, live-attenuated HMPV or recombinant viruses are a good choice for this population. Another vaccine strategy, prime-boost regimen, consists of vaccinating with gene-based/live vaccine first and then with a protein/particle-based vaccine [118]. Immunization of pregnant women can not only provide a passive antibody transfer to their children but also prevent a mother-to-child transmission of HMPV. Live vaccines are considered being too risky to be used in this population; therefore, subunit vaccines or VLPs with standard adjuvants should be considered. The effect of vaccination of adult populations with live vaccines can be hampered by the presence of anti-HMPV antibodies in the bloodstream or in the respiratory tract (RT). For the vaccination of elderly people, who had experienced multiple HMPV infections, subunit or VLPs with a potent adjuvant are recommended [118].
2. Vector-Based Chimeric Vaccines
One of the strategies aimed to circumvent the problem of incomplete immunity provided by natural HMPV infection is to express its protective antigens in the backbone of a more immunogenic virus. The increase in immunogenicity can also be provided by improved antigen expression by the vector, as demonstrated for a recombinant bovine/human PIV3 (rB/HPIV3) expressing either RSV-F or G proteins [60] and for human parainfluenza type 1 virus (HPIV1) expressing HMPV antigens [54]. HMPV is difficult to grow in cell culture; vectoring its antigens with the backbone of a better-replicating virus can facilitate vaccine development and manufacturing. For instance, rB/HPIV3 expressing RSV-F reaches 10–100-fold higher viral titers in vivo compared to wt RSV [60] and replicates more efficiently in the RT of AGMs than wt RSV or HMPV [119,120]. Expression of a foreign antigen in the backbone of another virus can also mitigate the problem of pre-existing immunity that often diminishes the effect of the vaccine. Using vectors of another host range can facilitate the immunization of seropositive individuals with no risk of causing disease, as it has been described for Newcastle Disease Virus (NDV) expressing RSV-F. The majority of live vaccines against RSV and HMPV up to date have been attenuated by a few codon changes in their genome. Therefore, using more stably attenuated backbone can render the attenuation more reliable, and decrease the risk of the reversion of an attenuated phenotype. The insertion of an additional gene can itself influence the virus’ replicative capacity, thus limiting its ability to spread and cause the disease, but also to create the risk of overattenuation. This subtle balance between attenuation and immunogenicity remains the major challenge of vaccine development.
In general, vector-based chimeric vaccines retain the advantages of live attenuated vaccines and they can be readily generated by reverse genetics. Most of the backbones used so far for vectoring pneumoviral antigens belong to the Paramyxoviridae family. These viruses are closely related, both phylogenetically and structurally, to pneumoviruses. This close relationship increases the chance of an efficient expression and integration of a pneumoviral protein in the background of a paramyxoviral vector. Among paramyxoviruses, the most widely used vaccine backbones are bovine parainfluenza virus type 3 (BPIV3) and its recombinant derivative rB/HPIV3, HPIV1, parainfluenza virus type 5 (PIV5), Newcastle Disease Virus (NDV), and Sendai virus (SeV). The reasons explaining this frequent use of paramyxoviruses as vaccine backbones are numerous: First, their genomes are well-characterized and complete genomic sequences of all known members of this family are easily accessible [121]. Second, their genomes are simple and organized in a modular way. Tandem alignment of the genes, of which the most are transcribed as separate mRNA products, facilitates genetic manipulations [122]. Third, most paramyxoviruses are able to replicate efficiently in cell lines certified for vaccine manufacture, such as Vero cells, which can facilitate vaccine development [122]. Fourth, as they replicate entirely in the cytoplasm and their replication cycle does not involve the integration into the host cell genome, their use is potentially safe [121]. Also, recombination events between mononegaviruses have not been observed in nature and they remain extremely rare in vitro, even in optimized conditions [123,124], indicating that there is a low probability of gene exchange between engineered vaccine viruses and the pathogens present in the environment. Fifth, the small probability of recombination also contributes to the stability of a genetic insert, providing a stable foreign gene expression system. Paramyxoviruses are able to stably express several exogenes simultaneously and the level of expression can be manipulated by changing the position of gene insertion [125]. Sixth, most paramyxoviruses infect their host via their RT, representing an easy and safe route of administration of the vaccine as well as for induction of both local and systemic immune responses. Last, but not least, there are many animal paramyxoviruses that are naturally attenuated in humans due to a host range restriction. Mutations that render these viruses harmless to people have been identified and characterized, thus making attenuation of different paramyxoviruses possible [122].
3. Recombinant Virus Engineering
3.1. Reverse Genetics
Negative strand viruses can be readily recovered from cell cultures by means of reverse genetics [126]. This technique makes it possible to engineer a fully functional virus starting from its genetic sequence. The genetic material in the form of cDNA can be easily modified according to the vaccine design. The technique is based on transfecting permissive cells with a plasmid coding for the viral genome and satellite plasmids coding for all the proteins necessary for the formation of a ribonucleoprotein complex (RNP) that initiates the transcription of viral genes (Figure 1). The proteins indispensable to form the RNP in paramyxoviruses and pneumoviruses are N, P, and L proteins [127,128,129], yet the addition of M2-1 protein facilitates the recovery HMPV from cDNA [130]. The elaboration of a polyvalent vaccine can be accomplished by either cloning additional genes into the plasmid coding for viral genome or replacing protective antigens of a vector with the ones of another pathogen.
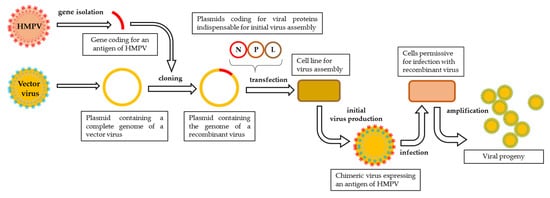
Figure 1. Schematic representation of reverse genetics pipeline. The exogene of interest is cloned into a plasmid containing a complete genome of a vector virus and then transfected, along with satellite plasmids coding for viral proteins indispensable to initiate viral assembly, into a cell line designed for initial virus production. The first progeny of the recombinant virus is harvested and propagated on a permissive cell line.
3.2. Genomic Organization of Paramyxoviridae and Pneumoviridae
The genomes of Paramyxoviridae and Pneumoviridae consist of a simple, nonsegmented, linear, single-stranded, 15,000–19,000-nucleotide-long negative RNA that contains 6–10 genes [121]. The most 3’-proximal region consists of a leader (le)—a 50-nucleotide-long promoter—and each gene is preceded and followed by a short (10–13 nucleotides) conserved sequence named gene start (GS) and gene end (GE). These sequences act like transcription control signals for viral RNA-dependent RNA-polymerase (vRNAP) and they guide the enzyme along the genome [131]. Single genes are separated from each other by short, non-coding intergenic regions and the order of the genes is usually conserved as 3’-Nucleoprotein (N), Phosphoprotein (P), Matrix Protein (M), Glycoprotein (G), Hemagglutinin-Neuraminidase (HN), or Large Polymerase subunit (L)–5’ with the presence and location of additional genes depending on the virus [132] (Figure 2).
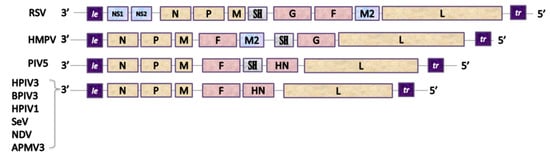
Figure 2. Genetic organization of Pneumoviruses and some of Paramyxoviruses used as viral vectors. Pneumoviridae: RSV: Respiratory Syncytial Virus (Orthopneumovirus), HMPV: Human Metapneumovirus (Metapneumovirus). Paramyxoviridae: PIV5: Parainfluenza type 5 virus (Rubulavirus), HPIV3: Human Parainfluenza type 3 virus, BPIV3: Bovine Parainfluenza type 3 virus, HPIV1: Human Parainfluenza type 1 virus and SeV: Sendai Virus (Respirovirus), NDV: Newcastle Disease Virus and APMV3: Avian Paramyxovirus type 3 (Avulavirus). Genes: le: leader, NS1 and NS2: accessory proteins of RSV, N: nucleoprotein, P: phosphoprotein, M: matrix protein, F: fusion protein, SH: small hydrophobic protein, G: attachment glycoprotein, HN: Haemagglutinin-Neuraminidase protein, M2: gene coding for M2-1 and M2-2 proteins, L: large polymerase subunit.
The 5’-end of a genome contains a trailer sequence (tr) of a variable length, ranging from 50 up to 707 nucleotides [121,133]. The genomic RNA of paramyxoviruses and pneumoviruses does not exist as an unbound RNA-particle: it is always assembled with numerous copies of N protein and forms a helicoidal nucleocapsid. In paramyxoviruses, each N protein molecule is associated with precisely six nucleotides, a feature that is believed to underlay the ‘’rule of six’’, i.e., the length of the genome of paramyxoviruses has to be a multiple of six for an effective viral replication [134,135]. The stringent adhesion to this rule is observed for SeV [136]; for other viruses, like PIV5, NDV, and HPIV3, it strongly increases the efficacy of replication [137,138,139]. The fact that it does not give any replicative advantage to RSV [140] might be explained by distinct differences in nucleocapsid structure of the two families [141]. Transcription of the viral genome is initiated at the 3’-end and a 3’-to 5’ expression gradient is observed. The complex of vRNAP sporadically fails to resume the synthesis of another distinct mRNA at each gene junction, which results in a gradual loss of transcription-efficacy along the genome [121].
3.3. Principles of Exogene Insertions
The main principles to be taken into consideration while engineering the genome of recombinant paramyxoviruses are the preferential adherence to the rule of six, a coherence of transcription control signals (GS/GE) used to drive the expression of a foreign antigen and exogene positioning in the transcription gradient. As it has been documented in numerous studies, the GS and GE signals of the vector virus can efficiently direct the expression of an exogenous protein [60]. Although GS and GE signals are often highly conserved along the viral genome, there might be variations in their sequences and transcription efficacy [142]. Flanking the sequence of GFP inserted into the 6th genome position of PIV5 with GS/GE specific for either 2nd or 7th gene junctions resulted in large differences in GFP expression levels (Figure 3) [143]. GS/GE characteristics for the 1st junction provided better expression levels than the ones originating from the 7th. It is therefore important to flank the exogene with potent transcription regulators. In accordance with the 3’-5’ transcription gradient, exogenes placed in more 3’-proximal position should be expressed better than 5’-proximal inserts, yet the tendency cannot be described as linear and some deviations are observed [144]. The adherence to this gradient might also be influenced by the type of attenuation of vector virus, as it has been demonstrated for HPIV1 bearing RSV-F protein [145]. The positions of an exogene in the viral genome are described in Figure 3 and the nomenclature used to label chimeric viruses in this work is explained in Figure 4.
Figure 3. Insert-positions in the paramyxovirus genomes.
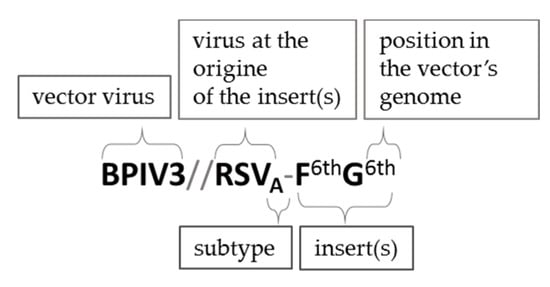
Figure 4. The nomenclature used in this work to describe chimeric viruses expressing additional antigens. The inserts are marked according to their 3’-5’ order of the vector’s genome. The subtype of the virus at the origin of the insert is marked if it is specified in the source. Although discussing different chimeric viruses based on the same vector that are mentioned in the same study, recombinant viruses can be also referred to as F1st, G2nd, etc. constructs. The nomenclature of vector backbones modified by additional mutations, protein swapping, etc. (i.e., rHPIV1-CΔ170, rHMPV-PA) was unchanged in relation to the source publication.
On one hand, a 3’-proximal insertion should provide the best level of expression; on the other hand, it can influence the level of transcription of all downstream genes, leading to a decrease in viral replication. RSV-F insertion into 1st position of rB/HPIV3 genome reduced the expression of downstream genes by 20–45% [146]; 2nd genomic position can provide a good expression, but it can also influence the N:P protein ratio, which plays a decisive role in the replicative capacity of paramyxoviruses, causing an additional attenuating effect [146]. Insertions in either 1st or 2nd genomic positions are the most privileged for PIV3-based vectors, with the 2nd position usually providing better virus recovery, higher viral replication, and better exogene expression [147]. For some vectors, namely, NDV and Avian paramyxovirus serotype 3 (APMV3), the 3rd position is the most advantageous, whereas the insertion at the 2nd one results in delayed viral replication and the largest reduction in virus recovery [148,149,150]. The 4th and the 5th positions are not frequently used, so as to not influence the expression of vector’s surface proteins (F and HN) [146], with the exception of studies on SeV bearing either RSV-F or HMPV-F at the 5th position [151,152,153].
The nature of the insert itself can also influence the vector’s biology. The rB/HPIV3//RSV-F1st virus showed an 8-fold reduced replication in vitro compared to rB/HPIV3, whereas the RSV-G1st insert did not influence viral replication [60]. This decrease in viral growth might have been due to excessive syncytia formation and increased cytopathology resulting from the expression of a second fusion protein. Another reason might have been the size of the insert–a bigger F protein might have influenced the replication more significantly than a smaller G protein. The size of an exogene can significantly change the replicative capacity of the virus, as it has been shown for HPIV3 vector bearing inserts of different sizes [154]. The level of integration of a foreign protein into a vector’s particle can also be a price to pay in the exchange for efficient expression of the exogene. Improved exogene integration into the vector’s backbone can be obtained by the substitution of the transmembrane domain (TM) and cytoplasmic tail (CT) of the inserts with their equivalents from the vector virus. As demonstrated for HPIV1 vector bearing RSV-F protein either in its wt or chimeric TMCT form, packaging of RSV-F(TMCT) was strongly improved, but the chimeric virus was overattenuated and not protective in hamsters [155].
This entry is adapted from the peer-reviewed paper 10.3390/pathogens9020135
This entry is offline, you can click here to edit this entry!
