As a rule, a single gene copy appears to be enough to provide development and life-cycle maintenance of diploid animals; however, a small cohort of genes exhibits a high sensitivity in case of decreased gene dosage. This phenomenon is known as haploinsufficiency, and it is associated with many developmental disorders in human
[47][48][49]. Comparing the evolution of flies and humans, one could assume that the Y-chromosomal genes, which have homologues on the X chromosome and do not directly contribute to the functioning of the male reproductive system, are relics that will disappear over time, as has apparently happened in flies. However, their maintenance can be determined by the peculiarities of the dosage compensation system in mammals. In male flies, the genes of the only X chromosome are overactivated in somatic tissues, eliminating the problem of haploinsufficiency and potentially lethal imbalance between the X and autosome transcriptional level in the two sexes. Therefore, in flies, X activation may eventually compensate for haploinsufficient homologous genes lost on Y, which is impossible in mammals. In contrast, in female mammals, inactivation of one of the two X chromosomes occurs. However, according to various estimates and in distinct types of human cells, 20–30% of genes of inactive X chromosome escape the inactivation
[50][51]. In mammals, haploinsufficient Y-chromosomal genes have X-chromosomal homologues that avoid inactivation during dosage compensation in females, which indicates the need for their expression on both sex chromosomes to ensure normal functions in the body. Thus, in males, these dosage-sensitive genes cannot disappear from the Y chromosome without negative consequences, and they can survive under selective pressure
[31]. This hypothesis is supported by the maintenance of functional X-Y gene pairs associated with housekeeping regulatory functions such as lysine demethylation, stem cell self-renewal, splicing, translation initiation, and deubiquitylation
[31][32][50][52]. Strict dosage requirements for sex-linked genes are demonstrated in the case of Turner syndrome (exhibiting X0 karyotype or mosaicism) and Klinefelter syndrome (XXY), since such genes have been haploinsufficient or overexpressed, respectively, in these karyotypes
[51]. Turner syndrome is a genetic condition caused by complete or partial loss of the second sex chromosome in human. Half the patients with Turner syndrome have the X0 karyotype (monosomy of the X chromosome), the other half exhibits mosaicism or a presence of the fragmented X or Y chromosomal material and other more complex karyotypes
[53][54]. Studies of manifestations of this syndrome indicate that the functions of the Y chromosome consist not only of ensuring the normal functioning of the male reproductive system. Due to the absence of the
SRY gene, which is the key to triggering male-type development, patients with this syndrome are exclusively female, with multiple body disorders: small size, rudimentary ovaries and infertility, pathologies of the cardiovascular system, autoimmune disorders, increased risk of developing diabetes, and cognitive impairment
[54]. Individuals with Klinefelter syndrome are infertile as a result of excess gene dosage of X escape genes, and abnormal meiotic pairing of the sex chromosomes. An atypical number of X or Y chromosomes (XXY, XXX, or X) contributes to spatial chromosome conformation changes and leads to disruption of DNA methylation patterns of autosomal genes, causing distinct disease phenotypes: mental illness, cancer, and disrupted fertility
[51].
Attempts to understand how these patterns are generated can be important not only for fundamental evolutionary biology, but also for biomedical challenges, since Y-chromosomal pathologies in humans differ from other genetic anomalies due to the unique nature of the Y chromosome. It is convenient to study the Y chromosome evolution in species with a rapid generation turnover, as in the Drosophila species.
Transcription of these extremely large genes and the processing of their transcripts, including splicing, has a high metabolic cost for cells. The study of genes possessing giant introns using the Drosophila model provides a useful insight into the problems of expression of such genes in humans and the pathologies associated with their improper transcription or splicing. Study of the molecular mechanisms of maintenance, transcription, and processing of Y-loop genes in Drosophila may improve understanding of the origin, selection, and regulation of genes with similar structures in other species.
4. Differential Expression of rDNA Loci of Drosophila Sex Chromosomes
4.1. Nucleolar Dominance as a Widespread Phenomenon
In eukaryotes, there is a known phenomenon of a different level of expression of genes represented in the genome by two or more alleles
[100][101]. Some of these alleles are expressed at a high level, while the expression of the rest of them is completely suppressed. One of the most striking examples of this phenomenon is the regulation of expression of loci encoding ribosomal RNA (rRNA), called nucleolar dominance. This phenomenon was initially discovered in interspecies hybrids of different taxonomic groups of animals
[102][103][104]. In interspecies hybrids between
D. melanogaster and
D. simulans, rRNA genes from the
D. melanogaster genome are predominantly expressed, while these genes from the
D. simulans genome are suppressed
[105]. However, later this process was also found within species. Nucleolar dominance has been observed in both the plant and animal kingdoms and is generally characterized by the dominant transcription of rRNA loci residing in only one chromosome
[106][107][108]. Among the reasons for this phenomenon, DNA cytosine methylation, histone methylation and deacetylation, small RNA functions, and different affinities between transcription factors and promoter sequences of ribosomal DNA (rDNA) loci are suggested
[109][110]. However, the exact mechanism of this phenomenon remains unclear to date.
rDNA loci are arranged as tandem repetitive rRNA gene clusters flanked by intergenic spacer sequences (IGSs)
[109][110]. They are transcribed by the RNA polymerase I machinery as long precursor transcripts, subsequently processed into mature ribosomal RNAs (18S, 5.8S, and 28S). The transcriptional activity of these loci is high and achieves about 50–60% of the total transcription of metabolically active cells
[102][111]. rRNAs are highly conserved, but the loci that encode them are among the most unstable elements of the genome due to their repetitive nature and high transcriptional activity. The number of copies of rRNA genes varies from 100 to 1000 in different organisms, and they are often distributed over many chromosomes, including ten loci in humans
[112]. In mice, about 200 rDNA repeats grouped into NORs (nucleolar organizer regions) are distributed among the short arms of six acrocentric chromosomes
[102]. rDNA can undergo intrachromatid recombination, which can lead to a loss of rDNA copies or to the formation of circular non-genomic units-extrachromosomal circular rDNAs (ERCs) accumulating in aging yeast cells
[113]. In addition, active transcription of rDNA occurs even during the S-phase of the cell cycle, which can cause a conflict between replication and transcription. Such conflicts lead to frequent double-strand breaks and rDNA instability
[114]. Given the multifactor nature of rDNA instability, the number of rDNA copies in the loci can vary greatly even within populations of the same species. For instance, in
D. melanogaster strains, the variation in the number of rDNA copies can reach a sixfold range
[115]. Similar differences in the number of rDNA copies have been shown for a number of other organisms, including mice and humans
[116]. Decreased rDNA copy number leads to so-called replicative senescence in yeast
[113][117][118]. Nevertheless, despite significant variations, there are mechanisms that maintain the number of rDNA copies both in populations and in the process of transmission to subsequent generations. Therefore, it is important to study the mechanisms of maintenance of rRNA gene copies at a level necessary for survival.
4.2. Y-Based Nucleolar Dominance in D. melanogaster Males
In
D. melanogaster, rDNA loci reside on the X and Y chromosomes, each containing from 100 to 360 copies of rDNA genes. The presence of rDNA loci on the sex chromosomes greatly simplifies their study compared to other model organisms in which such loci are numerous and distributed over a large number of chromosomes. In
D. melanogaster males, intraspecies epigenetic silencing of X chromosomal rDNA in males was shown by two research groups in 2012
[107][108]. However, these studies of nucleolar dominance were based on larval neuroblasts and total RNA preparations from adult flies. While rDNA gene transcripts on the X and Y chromosomes are highly homologous, some genes contain insertions of non-LTR retrotransposons
R1 and
R2 [119]. These retrotransposons are able to specifically recognize a 30 bp target sequence in the transcribed region of 28S rDNA and integrate into this region preventing correct transcription of the whole cistron. It has been suggested that the nucleolar dominance of Y-linked rDNA loci over those in the X chromosome is partly due to the different number of transposon insertions in the rDNA loci. X-chromosomal rDNA loci contain a higher proportion of genes disrupted by the transposons than Y-chromosomal ones
[120]. For instance, the wild-type X chromosome contains insertions in 80% of rDNA units out of 100, while the wild-type Y chromosome contains insertions in 60% of rDNA units out of 360
[107]. The Y chromosome, with approximately the same number of insertions in the rDNA loci, does not show complete dominance over the X chromosome, but provides codominance (some expression of rDNA from the X chromosome occurs). Thus, there must be other factors that, together with the abundance of insertions, contribute to the nucleolar dominance of the Y chromosome. Interestingly, all analyzed lines of
Drosophila whose Y chromosome did not exhibit complete dominance carried mutations in the genes of heterochromatin proteins
Su(var)2–5 (HP1) and
Su(var)3–9 (encoding histone H3K9 methyltransferase). However, the authors failed to establish an unambiguous relationship between mutations of heterochromatin protein genes and derepression of rDNA loci in the X chromosome
[107]. Previously, it has been shown that in the absence of H3K9 methylation (in the case of a
Su(var)3–9 mutation) and upon disruption of the siRNA pathway (a
dcr-2 mutation) disorganization of nucleoli, rDNA loci, and adjacent satellite DNAs were observed
[121]. Recently, nucleolar dominance was analyzed in detail in
dcr-2 and
Su(var)3–9 mutants. Males with the
dcr-2 mutation showed no significant change in Y chromosome dominance during fly development, while males with the
Su(var)3-9 mutation demonstrated a significant decrease in Y dominance in nervous tissue of larvae, imaginal discs, and germline stem cells (GSCs) of adult males
[122]. Thus, heterochromatin-mediated repression of rDNA loci may contribute to the mechanism that regulates of their activity.
A recent work describes the investigation of nucleolar dominance of Y-linked rDNA loci in male GSCs of
D. melanogaster [123]. Although in the testes of young males, most GSCs contain a single spherical nucleolus 2 µm in diameter; however, during aging, the proportion of GSCs with normal nucleolus morphology gradually decreased, while the proportion of GSCs with atypical morphology increased. Atypical nucleolus morphology was manifested both in the fragmentation of the nucleoli into several foci, or in the altered nucleolar shape. Authors found that only Y-linked rDNA loci are associated with the nucleolus with typical nucleolar morphology, while X-linked loci are not, regardless of age. These results suggest that Y-chromosomal rDNA is actively transcribed, while X rDNA is not, which is consistent with Y nucleolar dominance. However, atypical nucleolar morphology that occurs in GSCs with aging is associated with the activation of the silent rDNA loci on the X, and leads to the transcription of rDNA from two separate chromosomes, each of which forms a separate nucleolus. Activation of X rDNA probably compensates for the decrease in the number of active copies of Y-linked rDNA, which decreases during aging owing to conflicts between transcription and replication machineries causing rDNA instability
[114][123]. GSC nucleolar morphology and rDNA copy number reduction is heritable and passed to male offspring from old fathers. Strikingly, the authors find that nucleolar morphology can be recovered in individual GSCs of these F1 sons during the first 10 days after eclosion to restore normal Y dominant state of rDNA transcription. This indicates the existence of a mechanism to maintain the number of rDNA copies across generations. This mechanism may be adaptive for the following reasons: firstly, rRNA expression from only one chromosome can prevent rDNA deletions on the other chromosome, transcription from which is suppressed; secondly, having intact rDNA loci present may allow GSCs to prolong their lifespan
[123].
Recently, the SNP in situ hybridization method was used to analyze in detail the transcription of rDNA clusters from the X and Y chromosomes of
D. melanogaster [122]. Throughout
Drosophila male development, the codominance of X and Y rDNA loci changes to the dominance of those on the Y chromosome. The manifestation of Y dominance in most types of larval tissues, such as nervous tissue, imaginal discs, fat body, and enterocytes of the anterior part of the midgut, has been found. However, salivary glands containing a large number of polytenized chromosomes showed only a modest manifestation of Y dominance. In females, using the SNP method, the codominance between the two X chromosomes was confirmed.
Drosophila females with the XXY genotype also exhibit Y dominance, suggesting that the presence of the Y chromosome is necessary and sufficient for the dominance. However, in the ovaries of adult females, codominance is also observed in GSCs and cystoblasts, while in the nurse cells Y dominance is found. Thus, in the case of the presence of the Y chromosome, nucleolar dominance predominantly occurs independently of the sex of the cell. This leads to the assumption that the Y chromosome must contain a specific nucleotide sequence that allows dominance to occur. In general, the sequences in rDNA loci of the Y chromosome and/or its proximal regions may be essential for the nucleolar dominance. Moreover, these results do not exclude the possibility that the long arm of the Y chromosome is involved in this process
[122].
4.3. Non-Random Segregation of Sister Chromatids of Sex Chromosomes in Drosophila
Recent studies also point to the involvement of rDNA loci in the nonrandom segregation of sister chromatids during cell division. The intergenic spacer repeats are responsible for X-Y pairing in
D. melanogaster males
[124]. Sister chromatids are not always completely identical due to the presence of epigenetic marks that distinguish them. The asymmetric arrangement of these marks, as well as kinetochore proteins, can lead to selective recognition of chromatids. The divergence of such sister chromatids is apparently one of the causes of asymmetric cell division. Recent studies have shown that non-random sister chromatid segregation is mediated by rDNA loci
[125]. Earlier it was shown that non-random segregation in
Drosophila is characteristic of the sister chromatids of the X and Y chromosomes, but not the autosomes
[126]. Researchers used the
Drosophila strain carrying a deletion of 80% of heterochromatin in the wild-type X chromosome (
Df(1)bb158). In GSCs of such fly males, random segregation of X chromosome chromatids occurred, while the segregation of the Y chromosomes remained non-random, suggesting that a chromosomal element deleted in the
bb158 strain acts
in cis to mediate non-random sister chromatid segregation. This indicates that the genetic elements necessary for this phenomenon are present in the deleted heterochromatin. It can be concluded that this genetic element is located in the rDNA loci. A more detailed study revealed that IGS sequences and the protein that binds to these sequences, Indra, are responsible for the non-random segregation of sister chromatids
[125]. Unequal sister chromatid exchange can be proposed as a possible mechanism to increase rDNA copy number on one sister chromatid for restoration of the number of rDNA copies disrupted in GSCs by aging.
4.4. Differential Expression of rDNA Loci in Human
The phenomenon of nucleolar dominance appears to be common across multiple species. It has not been shown directly in humans, due to the distribution of rDNA loci in multiple autosomal regions making them difficult to analyze owing to their highly repetitive nature. However, only a part of rDNA loci is actively transcribed in human cell lines
[127][128][129], suggesting that these loci may also undergo activation or suppression. To date, the principles of silencing or activation of rDNA loci in humans remain unknown. Recently, with the aid of Oxford Nanopore sequencing technology, obvious differences between methylated and unmethylated rDNA gene arrays in human cells have been revealed. The ratio of transcriptionally active unmethylated copies versus methylated ones has been found to be lower in individuals with higher rDNA copy abundance, indicating a possible mechanism for maintenance of a stable number of active rDNA copies
[129].
5. Drosophila Y Chromosome in Studying of piRNA Biogenesis and Functioning of piRNA-Clusters
5.1. Brief Description of the piRNA System
The piRNA pathway provides both innate and adaptive immune system defense against the activity of transposable elements (TEs) leading to the protection of genome integrity in germinal tissues. It also participates in the maintenance of germline stem cells, regulation of protein-coding gene expression, the establishment of embryonic patterning (in Diptera), and transgenerational epigenetic inheritance
[130][131][132]. Small non-coding piRNAs 23-35 nt in length associated with proteins of the PIWI subfamily are present in animals from fungi to humans
[133][134][135]. piRNAs are generated from piRNA clusters, which are long precursors that are transcribed from heterochromatic regions containing fragments of transposons. piRNA precursors are processed to generate small piRNAs in perinuclear nuage granules. Mature piRNAs loaded into the proteins of the PIWI subfamily, forming piRNA-induced RNA silencing complexes (piRISCs). The generation of primary piRNAs triggers production of secondary piRNAs via an amplification system called the ping-pong cycle (
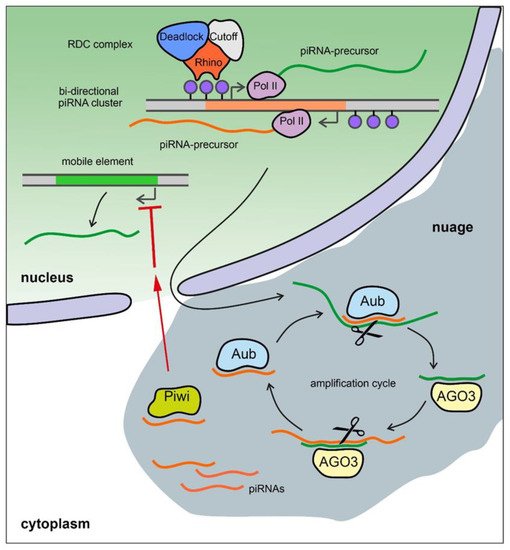
Figure 3)
[136]. The piRNA pathway is active, as a rule, in gonads and plays an essential role in fertility maintenance, preventing transposon activity and repressing harmful protein-coding genes. Transcripts of harmful genomic elements can be silenced post-transcriptionally via recognition and cleavage of complementary RNA-targets by piRISC complexes in the nuage. There is also a co-transcriptional repression mechanism, where recognition of nascent transcripts by piRISCs loaded with guide piRNAs leads to the establishment of heterochromatin in the corresponding genomic regions. Most of the known piRNA clusters in
Drosophila are bidirectional and transcribed with the participation of a specific Rhino–Deadlock–Cutoff (RDC) complex (
Figure 3)
[137][138][139]. Due to the Rhino chromodomain the RDC complex recognizes H3K9me3 histone modifications enriched in the chromatin of piRNA clusters and recruits to them the transcription initiation factor Moonshiner to promote non-canonical transcription. The RDC complex allows a skipping of transcription termination sites and inhibits splicing of piRNA precursor transcripts
[137][138][139]. In contrast, in mammals, the mechanisms of piRNA cluster expression seem to be indistinguishable from canonical Pol II transcription and include regular splicing and polyadenylation of the transcripts
[131]. However, the role of piRNAs may not be limited to germinal tissues. The involvement of the piRNA pathway in processes associated with disease, such as tumors of various etiologies, and aging in humans, has recently been shown
[140][141].
Figure 3. piRNA biogenesis in Drosophila germ cells. Bi-directional piRNA clusters are recognized by the RDC complex with the aid of histone modification H3K9me3 (blue dots) and are transcribed by Pol II machinery with the production of long unspliced transcripts of piRNA precursors. They are exported from the nucleus in the perinuclear nuage granules and are presumably cleaved by endonuclease Zucchini forming the 5′-end of the future piRNA (not shown). The cleaved transcripts are loaded into PIWI clade protein Aubergine (Aub) and then trimmed from the 3′-end by an unknown trimmer nuclease (not shown). Aub loaded with guide antisense piRNA recognizes and cleaves the complementary sense transcript producing the 5′-end of a new sense piRNA. The new piRNA is loaded into PIWI clade protein AGO3 and, in turn, performs cleavage of the complementary antisense transcript. This step generates a new antisense piRNA that is identical or very similar to the initiating piRNA (ping-pong amplification cycle). Piwi proteins loaded by antisense piRNAs translocate into the nucleus where they suppress transcription of TEs with complementary sequences by a co-transcriptional repression mechanism.
5.2. The Y Chromosome as a Major piRNA-Producing Genomic Region in the Fly Testes
The piRNA system in
D. melanogaster exhibits a strong sexual dimorphism. TE-mapping piRNAs are known as the most abundant class of piRNAs in the ovaries, whereas only about 40% of piRNAs map to TEs in the testes, and the largest cohort of piRNAs map to protein-coding genes
[7][142]. The genomic origin of most piRNAs between the two sexes is also different. In the testes of
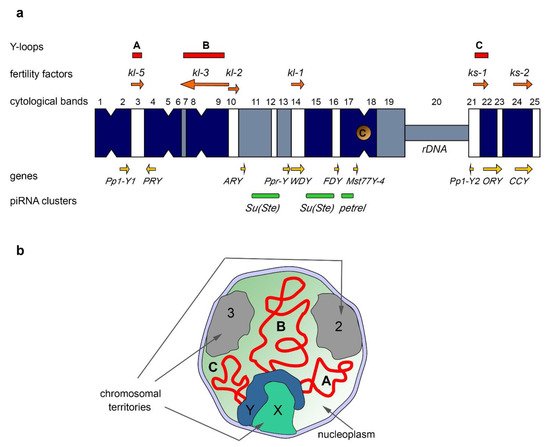
Drosophila, almost half of all piRNAs originate from the piRNA clusters located on the Y chromosome (
Figure 1a)
[7]. The largest number of piRNAs is generated from the Y-linked
Suppressor of Stellate (
Su(Ste)) repeats directed to silencing of the homologous tandem
Stellate genes residing on the X chromosome
[142][143][144]. The organization of the
Su(Ste) loci has been studied in detail. The number of
Su(Ste) repeats comprises more than 500 tandemly ordered copies residing in two cytolocations on the Y (
Figure 1a)
[2][4][7]. The size of a typical
Su(Ste) repeat is about 28 000 nt, consisting of three main parts: region homologous to the
Stellate genes, the AT-rich Y-specific region, and the insertion of transposon
hoppel (
1360) into the promoter.
Su(Ste) repeats are transcribed and processed to polyadenylated mRNAs; however, they contain numerous frameshift mutations owing to the presence of point mutations and deletions, and they are not translated
[145]. The insertion of the defective transposon
hoppel is responsible for the initiation of antisense transcription of
Su(Ste) repeats and their acquisition of piRNA cluster functions
[143].
Stellate derepression in the case of deletion of most of
Su(Ste) repeats or disruption of the piRNA system leads to the accumulation of needle-like protein aggregates in spermatocytes, disturbances of meiosis, and, as a result, a decrease in male fertility
[143][146]. The
Stellate/Su(Ste) system is species- and sex-specific for
D. melanogaster. Earlier, it was proposed that
Stellates are selfish genes involved in meiotic drive
[147]; however, no experimental pieces of evidence of this assumption have been found to date. Recently, it was is shown that
Stellate genes participate in male hybrid sterility of F1 progeny of crosses between
D. melanogaster females and
D. mauritiana males. The hybrid males possess maternal X-linked
Stellate genes, but their paternal Y chromosome does not contain
Su(Ste) repeats and the corresponding piRNAs are not generated. Derepression of
Stellates in the testes of hybrid males leads to a meiotic catastrophe and complete sterility
[142][146]. The contribution of the
Stellate/Su(Ste) system to reproductive isolation may explain the fixation and maintenance of this system in the
D. melanogaster genome. The acquisition of the
Stellate/Su(Ste) system by a part of the ancient fruit fly population could have been a causative factor of hybrid sterility in crosses of females with males that do not possess
Su(Ste) repeats on the Y. According to another speculation, the
Stellate/Su(Ste) system is similar to toxin–antitoxin systems, which are widespread in prokaryotes
[148].
Considering the amplification processes as an inherent property of the Y chromosomes, it can be assumed that some amplified repeats can be licensed as piRNA clusters during the evolution of a species. Given that not all piRNAs map to TEs, piRNAs produced by regions of the Y could exert sex-specific functions to regulate the expression of protein-coding genes besides
Stellate. If a gene has a positive effect on the processes occurring in the ovaries or other tissues, but, at the same time, reduces the efficiency of spermatogenesis, mechanisms for its testis-specific suppression can be developed, including piRNA-mediated silencing. The existence of a similar mechanism has been recently confirmed for the X-chromosomal
pirate/CG12717 gene, encoding a SUMO-isopeptidase. The Y chromosome of
D. melanogaster contains the
petrel locus (
Figure 1a), which is a source of multiple piRNAs highly complementary to
pirate, providing strong testis-specific silencing of this gene
[7]. However, the functional significance of the repression of
pirate in the testes remains unclear to date. It appears that both in the cases of the
Stellate/Su(Ste) and
pirate/petrel pairs, their current evolutionary relationships are initially based on parallel acquisition or co-amplification of homologous genes on the sex chromosomes. Note that in the
Drosophila testes, the expression and activity of the RDC complex is mainly limited to early stages of spermatogenesis, including GSCs and spermatogonial cells
[149]. The transcription of Y-linked
Su(Ste) and
petrel piRNA clusters takes place in primary spermatocytes, and it is independent from the RDC complex
[149]. The complex mosaic structure of
petrel repeats
[7] makes their further study as a functional piRNA cluster difficult. In the case of
Su(Ste), its sense transcription performs in the canonical manner from its own promoter, and the antisense transcription is initiated from several sites within the inserted transposon
hoppel [143][150], which makes it similar to mammalian piRNA clusters.
It has been assumed that in
Drosophila, maternal piRNAs, which are stored at the posterior pole of the oocyte during oogenesis, ensure the initiation of piRNA biogenesis from long piRNA precursors. However, this does not apply to Y chromosomal piRNA clusters due to the absence of the Y chromosome itself in females. According to a recent study, the determination of long RNAs as primary piRNA sources can also occur due to the recognition of specific
cis-regulatory 100-nt elements in piRNA precursor sequences, as in the case of the long non-coding RNA
flamenco and 3′ UTR of
tj mRNA in ovarian somatic cells
[151]. In many animals, including humans, the induction of germ cell precursors occurs from somatic pluripotent epiblast cells during embryogenesis, as a result of which all piRNA clusters are determined de novo
[132][135]. On the whole, the mechanism of determination of genomic regions as piRNA clusters is poorly resolved. Future studies of Y-chromosomal piRNA clusters in
Drosophila could allow us to elucidate these mechanisms both in
Drosophila and in mammals.
5.3. The Y Chromosome in Other Species as a Source of piRNAs
The suppression of genes harmful for spermatogenesis appears to be one of the main functions of piRNAs originating from the Y chromosome of
D. melanogaster. In mouse testes, novel polyadenylated non-coding RNAs called
Pirmy and
Pirmy-like transcribed from the long arm of the Y chromosome have recently been discovered
[152]. Multiple splice variants of
Pirmy encoded by a single locus have been identified experimentally; however, each exon of
Pirmy has been also found to be amplified in multiple copies on the Y chromosome. The 28
Pirmy-like RNA variants present various combinations of these exons that are distributed in multiple different loci on the mouse Y chromosome. Morphology- and sperm motility-related abnormalities have been found in two strains of Y-deleted mice with disrupted expression of
Pirmy and
Pirmy-like RNAs. The
Pirmy and
Pirmy-like RNAs serve as sources of piRNAs that are complementary to 5′- and 3′-UTRs of several autosomal genes, such as
FABP9,
Spink2,
superoxide dismutase (SOD), and
calreticulin, and also genes that presumably contribute to sex ratio maintenance in the progeny. The proteins expressed from these autosomal genes are up-regulated in the sperm of Y-deleted mice and appear to be responsible for the disruption of sperm morphology and motility
[152].
In
Bombyx mori, females are the heterogametic sex (ZW), and the W chromosome is heterochromatinized and consists almost entirely of transposon sequences. piRNA from the
Fem locus on the W chromosome functions as a suppressor of the
Masc gene, which regulates sex-specific splicing of the
doublesex (dsx) gene, which is necessary for sex determination in many insects
[153]. Thus, small piRNAs from the Y or W chromosome can potentially be involved in sex determination, the resolution of intragenomic conflicts, reproductive isolation, and the regulation of gene expression for ensuring spermatogenesis
[7][16][142][151][152][153][154].
Y-linked piRNA clusters and their functions in humans remain poorly understood
[155][156]. High-throughput sequencing of piRNAs from three human adult testis samples and subsequent data analysis have revealed 28 putative piRNA-cluster candidate regions on the Y
[157]. However, among them, only one uni-directional cluster with coordinates chrY:3231747-3235845 contains a significant number of mapped piRNAs (45.4 rpkm). This locus includes remnants of SINE, LINE, and LTR TEs, and has a highly homologous region of the same size on the X chromosome. The remaining 27 piRNA clusters predicted on the Y are rather small and remain unexplored. It should also be noted that due to the high level of heterochromatinization and a large number of repetitive elements, the human Y chromosome is not perfectly assembled, and data about piRNA clusters are not complete.
5.4. The Y Chromosome and TEs
Degeneration of the Y chromosome has been accompanied by the acquisition of transposable elements. The old Y chromosome of
D. melanogaster is strongly enriched with retrotransposons of different families. Earlier, in some laboratory strains of
D. melanogaster, active Gypsy elements restricted to the Y chromosome have been found
[158]. Using the latest genome assembly, it has been uncovered that
Dm412,
Gypsy,
Het-A,
Doc,
TART,
Mdg1,
Mdg3,
blood, and
FW TEs are prevalent on the Y chromosome of
D. melanogaster [4]. Y chromosomes of the
D. simulans clade are similarly enriched in retrotransposons relative to the rest part of the genomes; however, Y chromosomes from even closely related species accumulate distinct TE sets
[5]. Whether TE accumulation is coupled with beneficial developmental processes remains to be determined. The stability and non-random localization of TEs throughout the Y speaks in favor of their putative functional role in the host
[2][159]. Recently the organization of functional centromeres of
D. melanogaster has been resolved in detail, due to the mapping of CENP-A-occupied regions of all chromosomes. It has been found that CENP-A mapped DNA is mainly composed of retrotransposons and is often flanked and inserted by large blocks of satellite repeats
[160]. However, satellites are practically not found in the Y centromere, despite the fact that the whole Y chromosome is strongly enriched in simple tandem repeats
[4]. For instance, the Y centromere region consists of a tandem array of non-LTR mobile element
Jockey-3 [160], and its stability and active state is required for the maintenance of the centromere. The presence of telomere-specific
Het-A,
TAHRE, and
TART non-LTR retrotransposons in the pericentromeric region of the
D. melanogaster Y chromosome and in closely related species
[2][161] remains mysterious and can potentially be associated with their involvement in telomere maintenance
[162].
Recent data indicate that the Y chromosome of
D. melanogaster appears to be a cryptic library of active copies of TEs. The preference of TEs for insertion into the Y chromosome can potentially be beneficial for the host, providing immunity against active TEs in the testes by producing piRNAs. However, by analyzing testis single-cell sequencing data, Lawlor and colleagues found an unexpected burst of activity of TEs residing on the Y in early spermatocytes of
D. melanogaster [163]. This event occurs during a specific developmental period that coincides with the up-regulation of Y-chromosomal fertility factors and spermatocyte-specific transcription of many Y-linked genes
[73], as well as with a decreasing level of several components of the piRNA pathway. Piwi expression is not detected in primary spermatocytes
[164][165], nor is the RDC complex
[149]. As mentioned above, the RDC complex is responsible for transcription of bi-directional piRNA clusters providing the bulk of piRNAs for the suppression of TEs. Indeed, in primary spermatocytes, the piRNA machinery switches to the production of piRNAs from the non-canonical
Su(Ste) and
petrel clusters, ensuring the silencing of protein-coding genes
[7][142][143]. Note that moderate activation of TEs at this stage of spermatogenesis can, in a certain percentage of cases, lead to transposon insertions and mutations in functional genomic regions. Given that spermatogenesis is a highly redundant process, some TE activity leading to detrimental effects with a low frequency may be inconsequential. Eventually, some mutations can be adaptive for an individual and, will subsequently be fixed in the population. Thus, TE mobilization in this narrow developmental window leads to newly arising genetic variability important for the evolutionary adaptation of the population to changing environmental conditions.
6. Conclusions
Despite the fact that in some animal taxa Y chromosomes are completely absent, in most heterosexual eukaryotes Y chromosomes are maintained and perform various essential functions. These include sex determination, ensuring male fertility; correct segregation of meiotic chromosomes; regulation of the activity of rDNA repeats; epigenetic regulation of harmful elements, including TEs and protein-coding genes; contribution to interspecies hybrid sterility, and other responsibilities. The Y chromosome life cycle progresses through a series of stages common to many organisms, such as birth, accumulation of genes necessary for spermatogenesis, cessation of the recombination process, degeneration of the bulk of acquired sequences, and aging. Whereas in mammals the appearance of the Y chromosome has occurred once in a common ancestor of marsupials and placentals before their splitting, presumably 160-180 MYA, in Diptera and some fishes, Y chromosomes have arisen and disappeared several times during their evolutionary history. Studies of model organisms, Drosophila and mice, have fundamental significance for uncovering the shared properties of Y chromosomes of multiple species. The convergent nature of evolution of the Y chromosome allows researchers to consider that the data obtained in model organisms can be useful to a certain extent for the prediction of the human Y chromosome behavior in the future, as well as in understanding how the specific structure of this chromosome reflects its functions in normal and pathological conditions.