Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
When a group of three or more species are connected by hybridization, they form a syngameon.
- syngameon
- hybridization
- multispecies
1. Introduction
Interspecies hybridization is relatively common across taxa, with occurrence estimates of 25% in plants and 10% in animals [1], and is thought to be the cause of several major speciation events [2][3]. When a group of otherwise distinct species are connected by hybridization, they form a syngameon, a copulative community [4]. The discovery of the first natural syngameon in birch trees by Gunnarsson and the multidirectional hybridization of cultivated Saxifraga by Lloyd (in [4]) sparked the first serious investigations into hybridization itself, which continue to this day. From its inception, the word syngameon was used to collectively describe “a large number of different individuals [from different species], which are all apparently able to produce fertile offspring with one another; one very large pairing-community, one syngameon” [4]. However, shortly thereafter, Du Rietz [5] used the term to describe a polymorphic hybrid collective in which “species have got more or less lost”, precipitating an idea that syngameons were just a collection of taxonomic misfits unable to be classified. Later, other authors such as Cuenot [6], Grant [7], and Beaudry [8] reclaimed the word to mean “species linked by frequent or occasional hybridization, a hybridizing group of species”. Out of this definition was born the ecological species concept, which allowed for gene flow but separated species by their adaptations to particular niches, in an attempt to explain oak differentiation under gene flow. The now well-known oak syngameon was alternatively named a “multispecies” by Van Valen [9]. Templeton [10] began a pattern of syngameon misinterpretation when he conflated the term with a hybridizing pair, the aftermath of which can be seen in recent hybridization papers that use syngameon to describe any two species that hybridize (e.g., [11][12][13][14]. According to Boecklen [15], a syngameon is produced when a group of closely related species forms a complex set of hybrid combinations. The researchers recommend this use of syngameon to define a breeding system with a three or more multispecies network. The minimum number of three participating species is important in this context as it distinguishes the commonly studied hybridizing species pairs from a more complex and possibly synergistic interacting multispecies system [16]. It is important to note that the current definition does not distinguish between fertile and sterile hybrids, or diploid and polyploid systems, both of which could have varying impacts on the structure of the syngameon. However, the term seems to be restricted to naturally occurring hybrids, whereas the term coenospecies would refer to artificial hybrids (Glossary) [17].
2. How Do Syngameons Form and Collapse?
2.1. The Origin of Syngameons
For hybridization to occur, species must overcome any existing barriers to gene flow, which include pre- and postzygotic reproductive isolating mechanisms. Prezygotic barriers can consist of temporal, geographic, mechanical, behavioral, and genetic mechanisms, while postzygotic barriers can consist of hybrid sterility, hybrid inviability, and F2 breakdown [18]. While not always initially present, some of these barriers can form after the secondary contact of two lineages to prevent further hybridization and reinforce species boundaries [19]. Conversely, the initial lack of reproductive isolating mechanisms or the failure of reinforcement (Glossary) can lead to stable hybrid zones. Despite the numerous obstacles faced, hybridization is not rare [1][20]. So why, then, are syngameons so rarely reported? After all, syngameons are just hybridization events between three or more species. What makes adding this third species interaction so difficult? The answer may lie in how syngameons form and collapse.
2.2. The Birth and Death Hypotheses
2.2.1. The Rapid Radiation Hypothesis
The rapid radiation hypothesis [21][22] postulates that rapid radiations, or in other words, relatively quick and numerous speciation events, allowed for the repeated origins of hybrid lineages. Syngameons are able to form among the newly radiated species because reproductive isolating mechanisms have yet to develop (Figure 1). In turn, this gene flow can act as a catalyst for additional radiation by replenishing standing genetic variation, aiding in the consumption of unexploited resources and occupation of new niches [23]. Further, these hybrid lineages could speciate themselves, becoming hybrid species; however, this often requires the formation of reproductive barriers, which would exclude the newly formed species from the syngameon. Seehausen [22] used Heliconius butterflies to exemplify syngameons providing new adaptive traits and promoting ecological diversification. Using kiwifruit as an example, Liu [21] showed how syngameons developed during early radiation but later collapsed as species diversified into new ecological opportunities to reduce contact and competition (but see, ref. [24]). The classic examples of radiations, including Heliconius butterflies [25][26], Darwin’s finches [27][28], and African cichlid fish [29], showed a similar pattern as species numbers rose and underutilized resources became scarce, stabilizing selection occurred and species began to accumulate genomic incompatibilities [22]. While the concepts behind the hypothesis remain valid, it is difficult to prove if ancient syngameons formed during radiation events, as many participating species may have since gone extinct and the rapid timeline of diversification would make a transient hybridization event hard to detect. Current simplified methods to detect ancient hybridization (e.g., ABBA-BABA) can fail to distinguish population structure from actual introgression when population sizes are large [30], as could happen in syngameons and rapid radiations. However, with improving molecular and coalescent techniques, ghost lineages and ancient introgression events are becoming easier to trace [31][32][33].
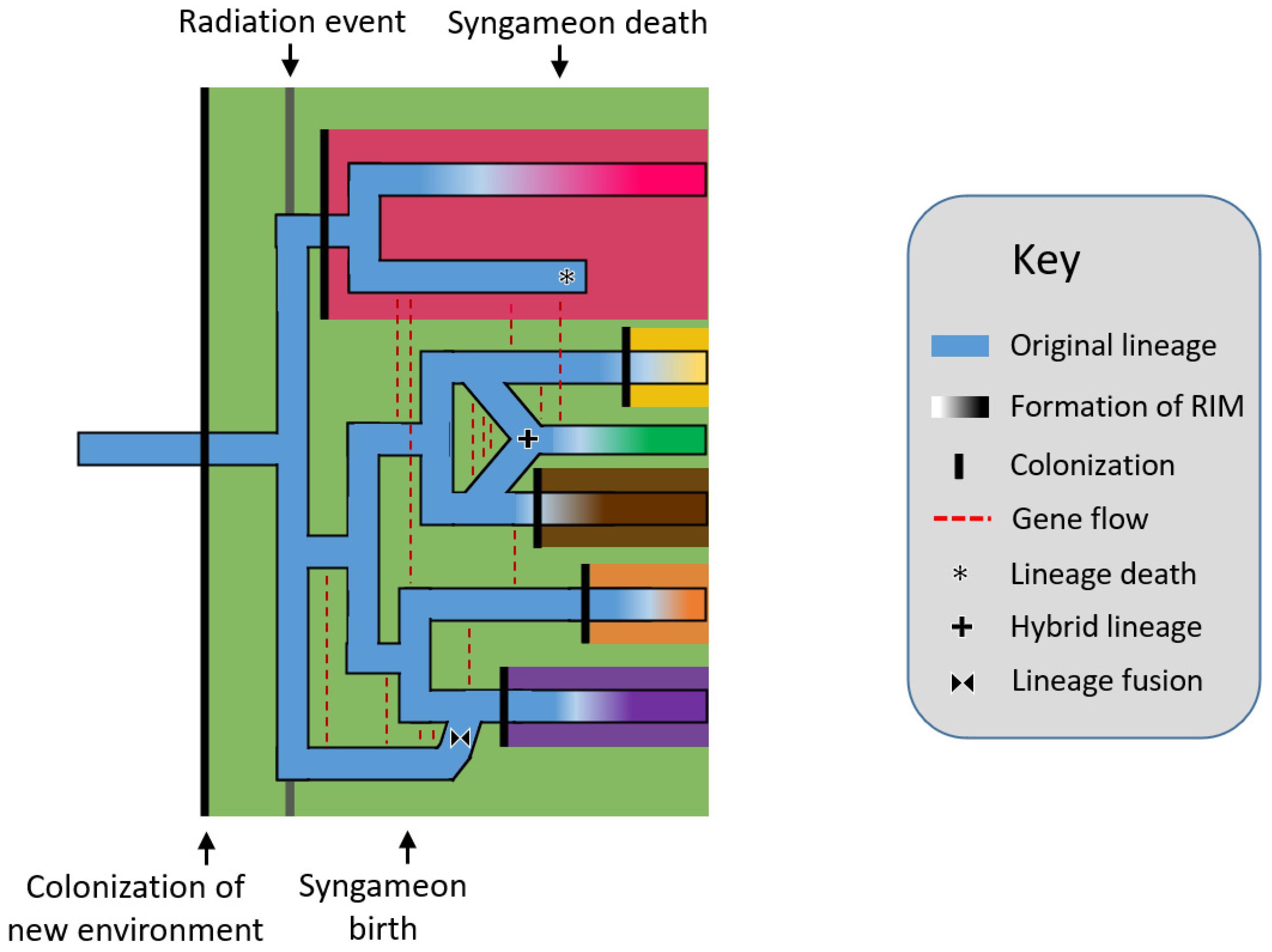
Figure 1. Rapid radiation hypothesis showing a lineage (blue horizontal line) colonizing a new environment (black vertical line), which eventually triggers a rapid radiation event. Speciation is followed by gene flow events (red dashed lines) which form a syngameon. The eventual collapse of the syngameon occurs when reproductive isolating barriers form among species, usually after the colonization of new environments, leaving two or no species with interspecific gene flow. Several potential outcomes are shown including hybrid speciation (plus symbol), extinction (asterisk), and fusion (bowtie symbol). RIM = reproductive isolation mechanism.
2.2.2. Surfing Syngameon Hypothesis
Rather than rapid radiation events causing syngameons, the surfing syngameon hypothesis [14][34] suggests syngameons that form during island colonization events can both cause and prevent rapid radiations. Distinct colonizing genotypes (referred to by Caujapé-Castells as morpho-species or incipient species) that were previously isolated on the mainland but are phylogenetically close enough to have gene flow can form a syngameon during a colonizing event of a new island (Figure 2A,B). The increase in genetic diversity would be enough to overcome selective pressures and founder effects, promoting the colonization of syngameon participants. Using species in the Canarian archipelago, Caujapé-Castells [34] indicated that this type of event could be detected through the level of endemic species, with low levels of endemism resulting from syngameon colonization and high levels from the formation of incompatibility barriers. In low-complexity islands, syngameons would stall evolutionary change due to high levels of gene flow homogenizing genomes, thus preventing rapid diversification. In high-complexity islands, syngameons would promote adaptations due to the high genetic diversity hybridization provides; therefore, resulting in rapid radiations and the eventual collapse of the syngameon (Figure 2C) due to competition and the formation of reproductive barriers [14]. Future studies should test the validity of this hypothesis beyond the Canary Islands and examine if it is broadly applicable to other allopatric dispersal scenarios such as the colonization of nearby niches or mosaic hybrid zones.
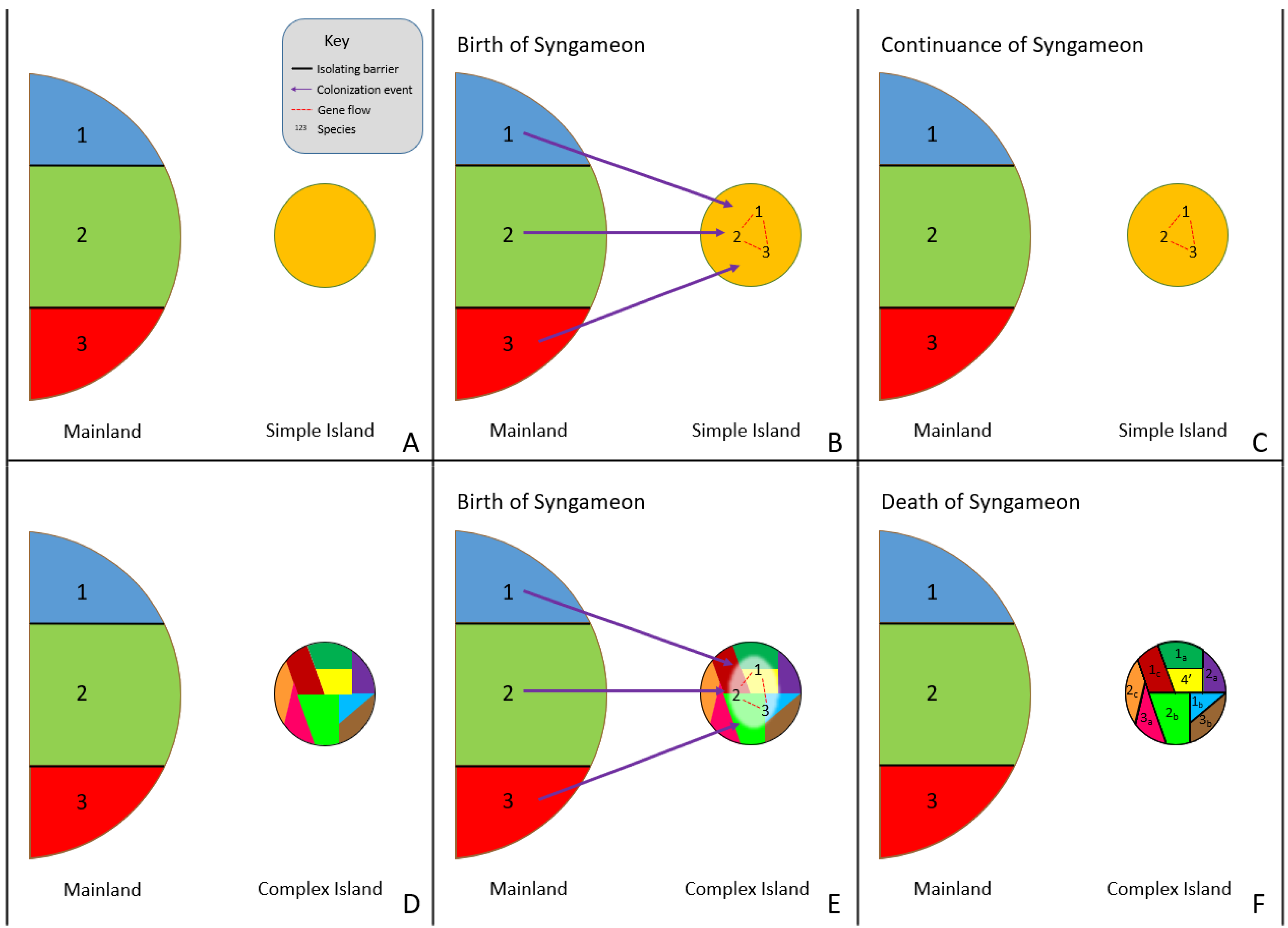
Figure 2. Surfing syngameon hypothesis, in which previously isolated species (1, 2, and 3) come into contact during the colonization of a low-complexity island (B) and high-complexity island (E), resulting in hybridization and the formation of a syngameon (B,E). The syngameon increases genetic diversity and reduces the effects of bottleneck events, resulting in the successful colonization of an island. If the island is open and uniform (A–C), with little to no ecological and geographical complexity (simple island), then evolutionary change is slowed down by syngameonic introgression/gene flow, resulting in homogenization of traits and the continuation of the syngameon (C). If the island is geographically and ecologically complex (D–F), then selection, adaptation, and competition eventually drive divergence and the formation of reproductive isolating barriers, resulting in the eventual collapse of the syngameon (F). Participation could even result in the creation of a new hybrid lineage (F, shown as lineage 4′).
2.2.3. Edge Range Hypothesis
Syngameons may form at the edges of species’ ranges, where multiple species can overlap in distribution (Figure 3A) [35]. Typically, range edges are seen as population sinks because the species is unable to adapt to the new, local environment beyond the current distribution boundary [36]. However, hybridization at a species’ range edge may facilitate survival by introducing locally adapted or novel traits through introgression [36]. Cronk and Suarez-Gonzalez [35] used a poplar syngameon to show how a tri-species interaction allows for the increased survival of hybrids at the edge of species boundaries. They also illustrated how as ranges expand and contract, these gene flow events could be episodic, explaining patterns of ancient introgression followed by divergence, then introgression again. However, Ottenburghs [37] pointed out that these “merging-and-diverging cycles” could result in the build-up of genetic divergence during allopatric phases, leading to lower levels of introgression during the following sympatric phase, eventually ending with a collapse of the syngameon. Additionally, Cronk and Suarez-Gonzalez [35] failed to consider the stability of syngameons at range edges because these interactions could lead to the formation of species barriers and thus the collapse of the syngameon, or even the formation of a new hybrid species with higher fitness than its parental species. Hybrid speciation could lead to the collapse of the syngameon and the possible extinction of the parent species via genetic swamping (Glossary) or hybrid superiority [38]. Moreover, there are examples of syngameons that do not form at the range edges, such as in Quercus in which some species overlap in wide ranges of the distribution [35]. As studies expand their scope beyond hybrid pairs to include more hybridizing species, range overlaps should be closely investigated in order to revisit this hypothesis under more scrutiny.
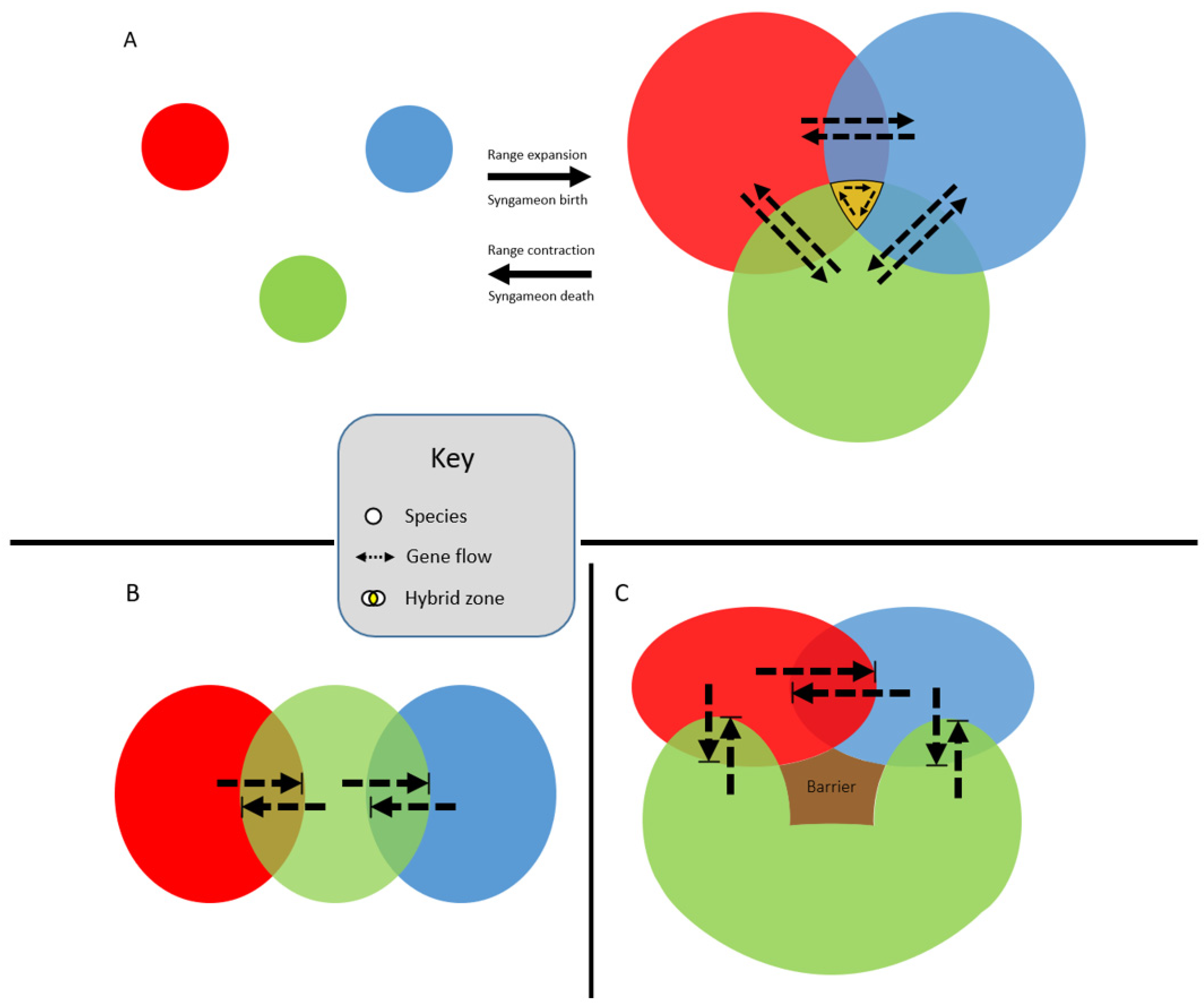
Figure 3. Edge-range hypothesis whereby the expansion and contraction of species’ ranges (A) over time makes gene flow within the syngameon episodic. This allows for the retention of species’ identities while still allowing for the exchange of adaptive alleles (dashed arrows). A caveat to the edge-range hypothesis is that all three species’ ranges rarely overlap (shown in gold). More probable scenarios are shown in (B,C), where species’ distributions overlap independently. While still technically syngameons, the scenarios represented in (B,C) may result in introgression not extending past the hybrid zones (bounded box), resulting in local admixture directly between hybrid pairs but no genes are shared indirectly through introgression via a third species.
2.2.4. Genomic Mutualist Hypothesis
Lastly, Cannon and Lerdau [39] hypothesized that species form syngameons by acting as genomic mutualists. In their scenario, multiple species remain partially interfertile with each other but experience divergent selection on portions of their genome, while low levels of neutral or adaptive gene flow occur in other parts of the genome. This creates a balance between purifying selection within species for specific phenotypes and diversifying selection among species for novel phenotypes. To avoid the negative consequences of extensive gene flow, species would develop a reduced but persistent capacity for interspecific mating, making periods of gene flow infrequent, episodic, and often unidirectional; however, in some systems, syngameons are multidirectional and often reciprocal gene flow occurs in different magnitudes [35][40][41]. These mating interactions are largely controlled by the quantity and quality of pollen or sperm, so interspecific gene flow would often be triggered by the decline of one species (Figure 4A,B), resulting in an overabundance of heterospecific gamete landing on the rare species (Figure 4B,C). This in turn allows for the rare species to avoid local extinction and inbreeding depression through the maintenance of diversity, a process called genetic rescue (Glossary; Figure 4A–C) [42][43]. However, demographic swamping (Glossary; Figure 4D), or genetic swamping (Figure 4E), where rare species are replaced by hybrids [38], is often used to counter this hypothesis as too much gamete swamping could instead result in the proliferation of hybrids and extinction of the rare species.
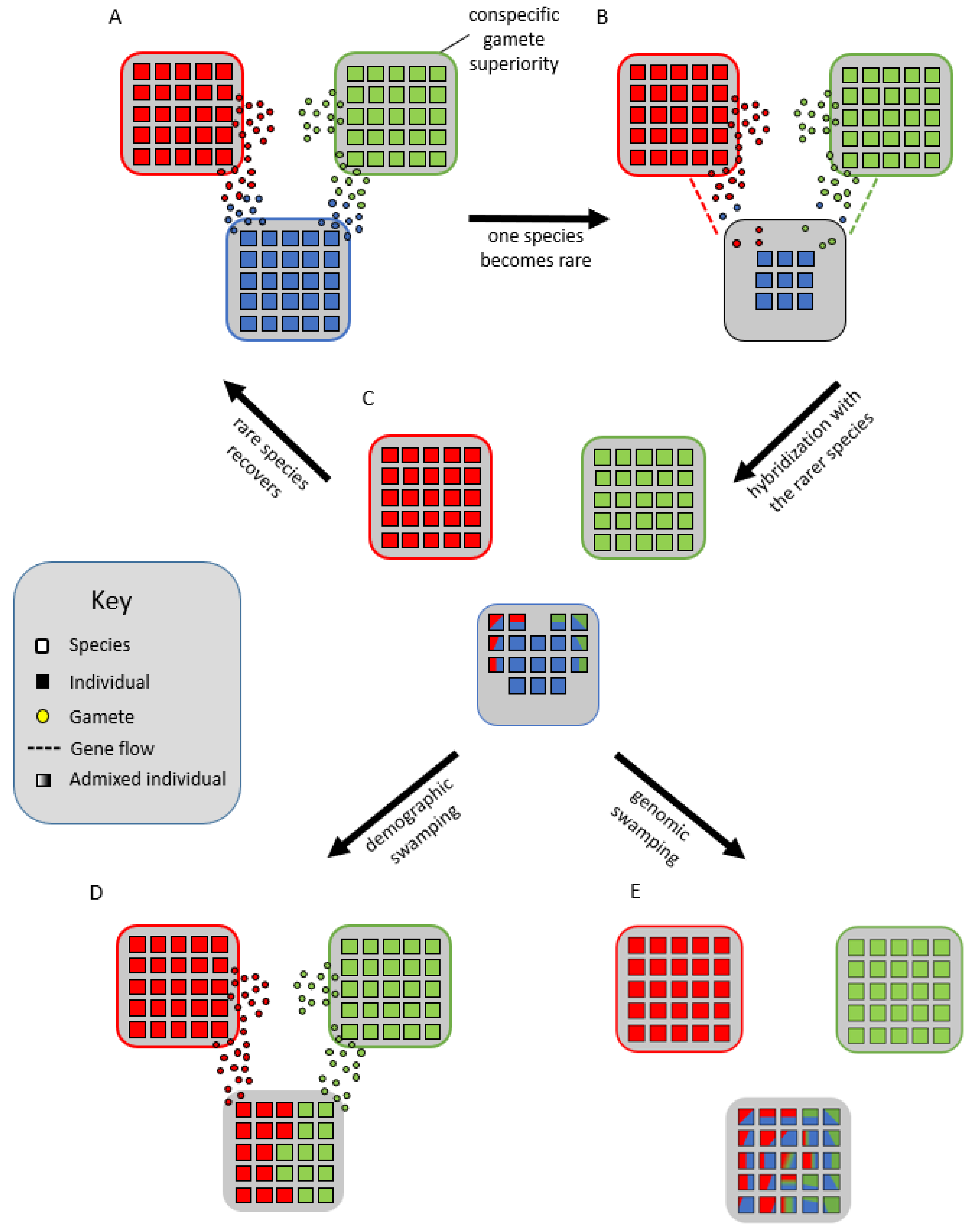
Figure 4. Genomic mutualist hypothesis in which there is a reproductive barrier favoring conspecific gametes (A) until one species becomes rare (B), wherein the gamete load from interspecific donors forces the rarer species to hybridize (C). This could lead to the rarer species benefiting from the increased genetic variation and effective population size, allowing it to overcome inbreeding depression and recover, a process known as genetic rescue (A). Alternatively, the rarity could lead to demographic swamping, where the rare species is replaced by the more abundant species through the purging of maladaptive hybrids (D), or genetic swamping, in which the rare species is replaced by admixed individuals (E).
2.3. Spatial Limitations
With the various ways syngameons are thought to form, it seems that there should be an abundance of syngameons. Perhaps the limiting factor is species distribution, meaning that, despite the number of species pairs with overlapping distributions reported to hybridize, the chances to have multiple hybridizable species with overlapping distributions is limited. In describing competition among highly diverse tropical tree communities, Cannon and Lerdau [39][44] found that direct spatial proximity with close relatives was infrequent, so even in complex ecological landscapes, the chances of overlapping with a congeneric species is low. Yet even if direct spatial overlap does not frequently occur, pollen and seed could still disperse into adjacent habitats and trigger syngameonic behavior. If, however, sympatry does occur, species in a syngameon could coexist and avoid competition by diversifying into microhabitats, as demonstrated by Schmitt et al. [45] in a Neotropical syngameon. Similarly, differing patterns of speciation may also play a role in limiting syngameon formation because allopatric species coming into secondary contact could be less likely to share a large enough portion of their range to overlap with more than one species. Further, the narrow hybrid zones that can result from secondary contact do not allow for the introgression of genes beyond the hybrid zone itself, which is usually at the edge of species’ ranges. Alternatively, F₁s could form but reproductive barriers could prevent any backcrossing with the parental species, thus introgression would not occur, as seen in Ligularia [46]. While many syngameon participants defined here hybridize with the same single species, genes are not necessarily introgressed across all species’ ranges, especially if the species hybridize at opposite ends of a range (Figure 3B,C). While this would still technically be considered a syngameon, the participants are not receiving all the benefits of the network-like structure of more sympatric syngameons. Cases of sympatric speciation may create more opportunities for multiple species to have overlapping distributions; however, these scenarios are rarer [47][48] and usually result in the formation of a reproductive isolating barrier [37][49][50], which would likely prevent any further hybridization. While the above syngameon formation hypotheses are not necessarily mutually exclusive, maintaining hybridization in multiple species at once can have compounding complications, with genetic swamping, lineage collapse, and the formation of reproductive barriers, all challenging the stability of a syngameon. If syngameons constantly fight to exist, then how are they maintained over time? The structure of known syngameons may shed light on this perplexing question.
3. How Are Syngameons Maintained over Evolutionary Time?
Most of the formation hypotheses above mention the episodic occurrences of gene flow within a syngameon and the limited amount of gene flow that must occur to stabilize the interactions. Yet most known examples of syngameons show extensive and constant gene flow among numerous participants. This discrepancy in theory and practice may be due to the varying hubs of introgression (Glossary), where some species contribute more genetic information than they receive and are connected to a large number of other species through gene flow [51]. The number of participating species can vary over geographical space and evolutionary time, with a single species (referred to as a hub) that has direct contact with multiple species, and as a result, genes passively introgress through the various pathways radiating from the hub (Figure 5). In these hub-based networks, if one pathway collapses, gene flow can still be maintained through the numerous other pathways connecting the species together, as long as there are no geographic or intrinsic barriers that act to contain alleles to one hybridizing species pair. However, if a hub disappears, that will likely have a larger effect on the entire network, but the extent of that effect is not currently known.
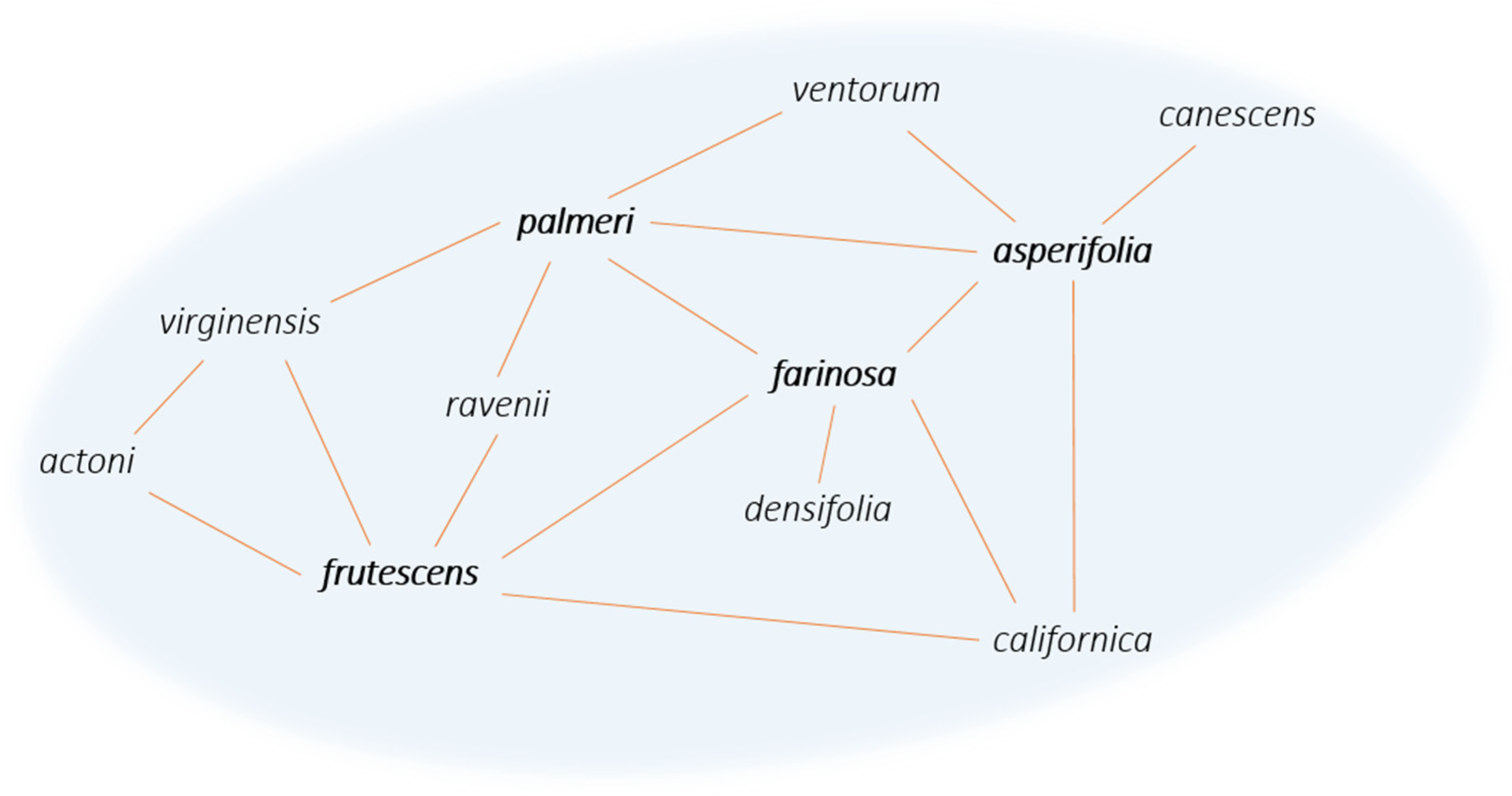
Figure 5. Network diagram of the Encelia syngameon [52]. Lines connecting species represent gene flow, with species such as E. frutescens, E. farinosa, E. palmeri, and E. asperifolia exemplifying hubs of introgression (in bold).
The direction and magnitude of these introgression pathways are rarely uniform. Boecklen [15] used simulations to test the structure of nine natural and four artificial syngameons (Glossary), finding that a majority exhibit a nonrandom structure, with a few species dominating the patterns of introgression. He concluded that geographically widespread species would have more opportunities to hybridize than restricted ones, with the Boechera syngameon demonstrating a positive relationship between geographic range and the number of mating combinations. The same is seen in North American white oaks, with the widespread Quercus alba mating with 11 out of 14 species in the syngameon [35]. It seems that the distribution of a species has a large impact on its ability to participate in the syngameon, with large contributors maintaining the structure of syngameons across geographical space. However, geographically widespread species may encounter more geographic and ecological barriers that could lead to population structuring or barriers to gene flow [53]. These could ultimately prevent the species, or at least certain populations, from participating in the syngameon or could result in reduced introgression beyond the hybrid zone. Additionally, the propensity to hybridize was unequal, even when species had equal opportunities to hybridize [15]. This suggests that there are other factors beyond range that affect the direction and magnitude of introgression within a syngameon. Genetic distance (Glossary) may be the largest of these factors, with closely related species hybridizing more readily than distant ones [15][54]. This would mean that the structure of syngameons is partially dependent on the relatedness of the species participating. Hypothetically, as time passes, species would become more distinct, compromising the structure and putting the maintenance of the syngameon at risk. However, the occasional gene flow events among syngameon members would counteract divergence and keep genomic distance smaller.
The evolutionary advantages and disadvantages of interspecific gene flow are well understood [3][18][55][56], but it is not known if these consequences are the same in these multispecies networks. Cannon and Petit [16] suggest that syngameons have synergistic properties, with network-like benefits that total more than just the sum of individual species pairs. Schmitt et al. [45][57] suggest that two contrasting evolutionary pressures are constantly acting on a syngameon, one at the species level to maximize individual species’ fitness and reduce competition among species, and one at the syngameon level to increase genus survival and maximize population size. In syngameons, adaptive introgression can maintain hybrid zones through the sharing of beneficial alleles [58]. Natural selection plays a role in maintaining the poplar syngameon when adaptive alleles are episodically exchanged across species boundaries [41]. Syngameons can also have increased heterozygosity, while maintaining partial infertility among species [59]. In the Fabaceae family, this partial infertility prevents genomes from fully merging, while still allowing gene flow to increase heterozygosity [59]. Levi et al. [60] suggested syngameons could help fuel the Red Queen arms race (Glossary) in tropical trees by increasing heterozygosity and introducing novel phenotypes. These beneficial outcomes of gene flow help maintain syngameons and can counter the negative complications that arise with hybridization. While the current definition does not differentiate between fertile hybrids that can backcross with their parental species and sterile hybrids that would prevent introgression, the hypothesized synergistic effects would likely only exist in the former situation where adaptive traits can be passed across species barriers. Further, the creation of infertile hybrids would more likely result in demographic swamping (Glossary; Figure 4D) and be detrimental to the syngameon as a whole.
There are many ways that syngameons can remain stable over long periods of evolutionary time including uneven participation, geographic distribution, genetic distance, and direction of gene flow within a syngameon. These factors can allow gene flow to occur episodically or in minute amounts, preserving the beneficial aspects of hybridization while avoiding the detrimental ones. A common misconception with hybridization is that it is ephemeral and only a stopping point on the way to reproductive isolation [61]. While time since divergence is positively correlated with the strength of reproductive barriers [62], classic two-species hybrid zones can be stable over evolutionary time through the balance between selection and dispersal [63], so it is reasonable that multispecies hybrid zones, while more complex, can be maintained in the same way. Without strong selection for the formation of reproduction barriers and with occasional gene flow partially homogenizing genomes, isolating barriers may take even longer to form within a syngameon, if at all.
This entry is adapted from the peer-reviewed paper 10.3390/plants11070895
References
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237.
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588.
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246.
- Lotsy, J. Species or linneon. Genetica 1925, 7, 487–506.
- Du Rietz, G.E. The Fundamental Units of Biological Taxonomy; Svensk Botaniska Foreningen: Uppsala, Sweden, 1930.
- Cuénot, L. L’Evolution Biologique: Les Faits, Les Incertitudes; Masson et Cie: Paris, France, 1951; p. 594.
- Grant, V. The plant species in theory and practice. In The Species Problem; Mayr, E., Ed.; American Association for the Advancement of Science: Washington, DC, USA, 1957; pp. 39–80.
- Beaudry, J.R. The species concept: Its evolution and present status. Rev. Can. Biol. 1960, 19, 219–240.
- Van Valen, L. Ecological species, multispecies, and oaks. Taxon 1976, 25, 233–239.
- Templeton, A.R. The meaning of species and speciation: A genetic perspective. In Speciation and Its Consequences; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 3–27.
- Arduino, P.; Verra, F.; Cianchi, R.; Rossi, W.; Corrias, B.; Bullini, L. Genetic variation and natural hybridization between Orchis laxiflora and Orchis palustris (Orchidaceae). Plant Syst. Evol. 1996, 202, 87–109.
- Holliday, T.W. Neanderthals and modern humans: An example of a mammalian syngameon? In Neanderthals Revisited: New Approaches and Perspectives; Springer: Berlin/Heidelberg, Germany, 2006; pp. 281–297.
- Barker, M.J. The empirical inadequacy of species cohesion by gene flow. Philos. Sci. 2007, 74, 654–665.
- Caujapé-Castells, J.; García-Verdugo, C.; Marrero-Rodríguez, Á.; Fernández-Palacios, J.M.; Crawford, D.J.; Mort, M.E. Island ontogenies, syngameons, and the origins and evolution of genetic diversity in the Canarian endemic flora. Perspect. Plant Ecol. Evol. Syst. 2017, 27, 9–22.
- Boecklen, W.J. Topology of syngameons. Ecol. Evol. 2017, 7, 10486–10491.
- Cannon, C.H.; Petit, R.J. The oak syngameon: More than the sum of its parts. New Phytol. 2020, 226, 978–983.
- Turesson, G. The species and the variety as ecological units. Hereditas 1922, 3, 100–113.
- Widmer, A.; Lexer, C.; Cozzolino, S. Evolution of reproductive isolation in plants. Heredity 2009, 102, 31–38.
- Garner, A.G.; Goulet, B.E.; Farnitano, M.C.; Molina-Henao, Y.F.; Hopkins, R. Genomic signatures of reinforcement. Genes 2018, 9, 191.
- Moran, B.M.; Payne, C.; Langdon, Q.; Powell, D.L.; Brandvain, Y.; Schumer, M. The genomic consequences of hybridization. eLife 2021, 10, e69016.
- Liu, Y.; Li, D.; Zhang, Q.; Song, C.; Zhong, C.; Zhang, X.; Wang, Y.; Yao, X.; Wang, Z.; Zeng, S. Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 2017, 215, 877–890.
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207.
- Kagawa, K.; Seehausen, O. The propagation of admixture-derived adaptive radiation potential. Proc. R. Soc. B 2020, 287, 20200941.
- Yang, R.; Folk, R.; Zhang, N.; Gong, X. Homoploid hybridization of plants in the Hengduan mountains region. Ecol. Evol. 2019, 9, 8399–8410.
- Beltrán, M.; Jiggins, C.D.; Bull, V.; Linares, M.; Mallet, J.; McMillan, W.O.; Bermingham, E. Phylogenetic discordance at the species boundary: Comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol. Biol. Evol. 2002, 19, 2176–2190.
- Gilbert, L.E. Adaptive Novelty through Introgression in Heliconius Wing Patterns: Evidence for Shared Genetic “Tool Box” from Synthetic Hybrid Zones and a Theory of Diversification; University of Chicago Press: Chicago, IL, USA, 2003; pp. 281–318.
- Grant, B.R.; Grant, P.R. High survival of Darwin’s finch hybrids: Effects of beak morphology and diets. Ecology 1996, 77, 500–509.
- Freeland, J.R.; Boag, P.T. The mitochondrial and nuclear genetic homogeneity of the phenotypically diverse Darwin’s ground finches. Evolution 1999, 53, 1553–1563.
- Seehausen, O.; Alphen, J.J.V.; Witte, F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 1997, 277, 1808–1811.
- Martin, S.H.; Davey, J.W.; Jiggins, C.D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015, 32, 244–257.
- Twyford, A.D.; Ennos, R. Next-generation hybridization and introgression. Heredity 2012, 108, 179–189.
- Hey, J.; Chung, Y.; Sethuraman, A.; Lachance, J.; Tishkoff, S.; Sousa, V.C.; Wang, Y. Phylogeny estimation by integration over isolation with migration models. Mol. Biol. Evol. 2018, 35, 2805–2818.
- Ottenburghs, J. Ghost introgression: Spooky gene flow in the distant past. Bioessays 2020, 42, 2000012.
- Caujapé-Castells, J.; Bramwell, D. Jesters, red queens, boomerangs and surfers: A molecular outlook on the diversity of the Canarian endemic flora. In The Biology of Island Floras; Bramwell, D., Caujapé-Castells, J., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 284–324.
- Cronk, Q.; Suarez-Gonzalez, A. The role of interspecific hybridization in adaptive potential at range margins. Mol. Ecol. 2018, 27, 4653–4656.
- Pfennig, K.S.; Kelly, A.L.; Pierce, A.A. Hybridization as a facilitator of species range expansion. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161329.
- Ottenburghs, J. The genic view of hybridization in the Anthropocene. Evol. Appl. 2021, 14, 2342–2360.
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G. Hybridization and extinction. Evol. Appl. 2016, 9, 892–908.
- Cannon, C.H.; Lerdau, M. Variable mating behaviors and the maintenance of tropical biodiversity. Front. Genet. 2015, 6, 183.
- Buck, R.; Hyasat, S.; Hossfeld, A.; Flores-Rentería, L. Patterns of hybridization and cryptic introgression among one-and four-needled pinyon pines. Ann. Bot. 2020, 126, 401–411.
- Chhatre, V.E.; Evans, L.M.; DiFazio, S.P.; Keller, S.R. Adaptive introgression and maintenance of a trispecies hybrid complex in range-edge populations of Populus. Mol. Ecol. 2018, 27, 4820–4838.
- Ingvarsson, P.K. Restoration of genetic variation lost—The genetic rescue hypothesis. Trends Ecol. Evol. 2001, 16, 62–63.
- Carlson, S.M.; Cunningham, C.J.; Westley, P.A. Evolutionary rescue in a changing world. Trends Ecol. Evol. 2014, 29, 521–530.
- Cannon, C.H.; Lerdau, M.T. Demography and destiny: The syngameon in hyperdiverse systems. Proc. Natl. Acad. Sci. USA 2019, 116, 8105.
- Schmitt, S.; Tysklind, N.; Hérault, B.; Heuertz, M. Topography drives microgeographic adaptations of closely related species in two tropical tree species complexes. Mol. Ecol. 2021, 30, 5080–5093.
- Zhang, N.-N.; Yu, J.-J.; Wang, Y.-H.; Gong, X. Molecular evidence for asymmetric hybridization in three closely related sympatric species. AoB Plants 2018, 10, ply011.
- Barraclough, T.G.; Vogler, A.P. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 2000, 155, 419–434.
- Foote, A.D. Sympatric speciation in the genomic era. Trends Ecol. Evol. 2018, 33, 85–95.
- Smith, J.M. Sympatric speciation. Am. Nat. 1966, 100, 637–650.
- Schliewen, U.K.; Tautz, D.; Pääbo, S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 1994, 368, 629–632.
- Ladner, J.T. Hidden Diversity in Corals and Their Endosymbionts. Ph.D. Thesis, Stanford University, Stanford, CA, USA, March 2012.
- Singhal, S.; Roddy, A.B.; DiVittorio, C.; Sanchez-Amaya, A.; Henriquez, C.L.; Brodersen, C.R.; Fehlberg, S.; Zapata, F. Diversification, disparification and hybridization in the desert shrubs Encelia. New Phytol. 2021, 230, 1228–1241.
- Nikolakis, Z.L.; Orton, R.; Crother, B.I. Fine scale population structure and extensive gene flow within an Eastern Nearctic snake complex (Pituophis melanoleucus). bioRxiv 2021, 51, 133–146.
- Larcombe, M.J.; Holland, B.; Steane, D.A.; Jones, R.C.; Nicolle, D.; Vaillancourt, R.E.; Potts, B.M. Patterns of reproductive isolation in Eucalyptus—A phylogenetic perspective. Mol. Biol. Evol. 2015, 32, 1833–1846.
- Rieseberg, L.H.; Wendel, J.F. Introgression and its consequences in plants. Hybrid Zones Evol. Process 1993, 70, 109.
- Tallmon, D.A.; Luikart, G.; Waples, R.S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004, 19, 489–496.
- Schmitt, S.; Hérault, B.; Ducouret, É.; Baranger, A.; Tysklind, N.; Heuertz, M.; Marcon, É.; Cazal, S.O.; Derroire, G. Topography consistently drives intra-and inter-specific leaf trait variation within tree species complexes in a Neotropical forest. Oikos 2020, 129, 1521–1530.
- Suarez-Gonzalez, A.; Lexer, C.; Cronk, Q.C. Adaptive introgression: A plant perspective. Biol. Lett. 2018, 14, 20170688.
- Chen, S.-C.; Cannon, C.H.; Kua, C.-S.; Liu, J.-J.; Galbraith, D.W. Genome size variation in the Fagaceae and its implications for trees. Tree Genet. Genomes 2014, 10, 977–988.
- Levi, T.; Barfield, M.; Holt, R.D.; Terborgh, J. Reply to Cannon and Lerdau: Maintenance of tropical forest tree diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 8106.
- Cannon, C. Is speciation an inexorable march to reproductive isolation? Mol. Ecol. 2021, 30, 4349–4352.
- Rothfels, C.J.; Johnson, A.K.; Hovenkamp, P.H.; Swofford, D.L.; Roskam, H.C.; Fraser-Jenkins, C.R.; Windham, M.D.; Pryer, K.M. Natural hybridization between genera that diverged from each other approximately 60 million years ago. Am. Nat. 2015, 185, 433–442.
- Barton, N.H. The dynamics of hybrid zones. Heredity 1979, 43, 341–359.
This entry is offline, you can click here to edit this entry!
