Clinically significant atherosclerosis of the coronary arteries, known as coronary artery disease (CAD), is an endemic condition that is associated with significant morbidity and mortality. The introduction of artificial intelligence (AI) and machine learning over the last two decades has unlocked new dimensions in the field of cardiovascular medicine. From automatic interpretations of heart rhythm disorders via smartwatches, to assisting in complex decision-making, AI has quickly expanded its realms in medicine and has demonstrated itself as a promising tool in helping clinicians guide treatment decisions. Understanding complex genetic interactions and developing clinical risk prediction models, advanced cardiac imaging, and improving mortality outcomes are just a few areas where AI has been applied in the domain of coronary artery disease.
- artificial intelligence
- coronary artery disease
- major adverse cardiovascular events
1. Integration of Genetics and AI in Cardiovascular Diseases
Over the last two decades, the emergence of technologies able to measure biological processes at a large scale have resulted in an enormous influx of data. For instance, the completion of the Human Genome Project has paved the way to design single-nucleotide polymorphism (SNP) and mRNA microarrays, which can broadly test for millions of genetic variants in a simple point-of-care test. This has paved the way for the emergence of modern data-driven sciences such as genomics and other “omics” [1]. Genome-wide association studies (GWASs) operate by simultaneous comparison of millions of SNPs between diseased individuals and disease-free controls to detect a statistically significant association between an SNP locus and a particular condition [1]. Machine learning (ML) and particularly deep learning (DL) algorithms are inherently designed to extract patterns and associations from large-scale data, including clinical and genomic data. Given the complexity and multifaceted nature of cardiovascular diseases in general, and CAD in particular, an approach that integrates all these factors into a risk-stratification model would be expected to better predict incident events than existent models [2].
Multiple studies have emphasized the role of ML in identifying genetic variants and expression patterns associated with CAD from mRNA arrays using differential expression analysis and protein–protein interaction networks [3][4]. For example, Zhang et al. used ML to perform differential expression analysis on mRNA profiles from CAD patients and healthy controls to identify a set of differentially expressed genes between the two groups, then built a network representation of functional protein–protein interaction. The top 20 genes in the network were identified using a maximal clique centrality (MCC) algorithm. Finally, to test the performance, a logistic regression model was built using the top five predictor genes to classify individuals into the presence or absence of CAD. The model achieved an AUC of 0.9295 and 0.8674 in the training and internal validation sets respectively [5].
Dogan et al. built an ensemble model of eight random-forest (RF) classifiers to predict the risk of symptomatic CAD using genetic and epigenetic variables along with clinical risk factors. The model was trained on a cohort derived from the Framingham heart study (n = 1545) and utilized variables derived from genome-wide array chips to extract epigenetic (DNA methylation loci) and genetic (SNP) profiles. The initial number of available variables were 876,014 SNP and DNA methylation (CpG) loci, which required multiple reduction steps, ending up with 4 CpG and 2 SNP predictors fed into the model along with age and gender. The model predicted symptomatic CAD with an accuracy, sensitivity, and specificity of 0.78, 0.75, and 0.80, respectively, in the internal validation cohort (n = 142).
Finally, the coronary artery calcium (CAC) score, calculated using the Agatston method on noncontrast ECG-gated cardiac computed tomography, is an established strong predictor of major adverse cardiovascular events in asymptomatic individuals. Genomic studies have previously focused on identifying genetic loci linked to CAC [6][7]. Oguz et al. suggested the use of ML algorithms to predict advanced CAC from SNP arrays and clinical variables. They identified a set of SNPs that ranked the highest in predictive importance and correlated with advanced CAC scores, defined as the 89th–99th percentile CAC scores in the derivation and replication cohorts, and trained different RF models to predict advanced CAC scores using clinical and genetic variables.
2. Risk Prediction Models and Imaging Modalities for Estimating Pretest Probability of CAD
When available, a CAC score has been shown to add to the PTP of CAD, with a CAC score of zero identifying low-risk patients who might not need additional testing [17][18]. ML models, combining clinical and imaging parameters, have been shown to have higher predictive power than traditional risk scores when predicting the PTP of obstructive CAD [19][20].
Various ML algorithms based on stress imaging, particularly single-photon emission computed tomography (SPECT), have been devised to facilitate the prediction of CAD. These models combined the clinical and demographic characteristics with the quantitative variables, as evaluated via SPECT to better predict CAD compared with the visual interpretation or quantitative variables alone [21][22][23][24][25][26].
3. Artificial Intelligence in Management of CAD in the Emergency Department
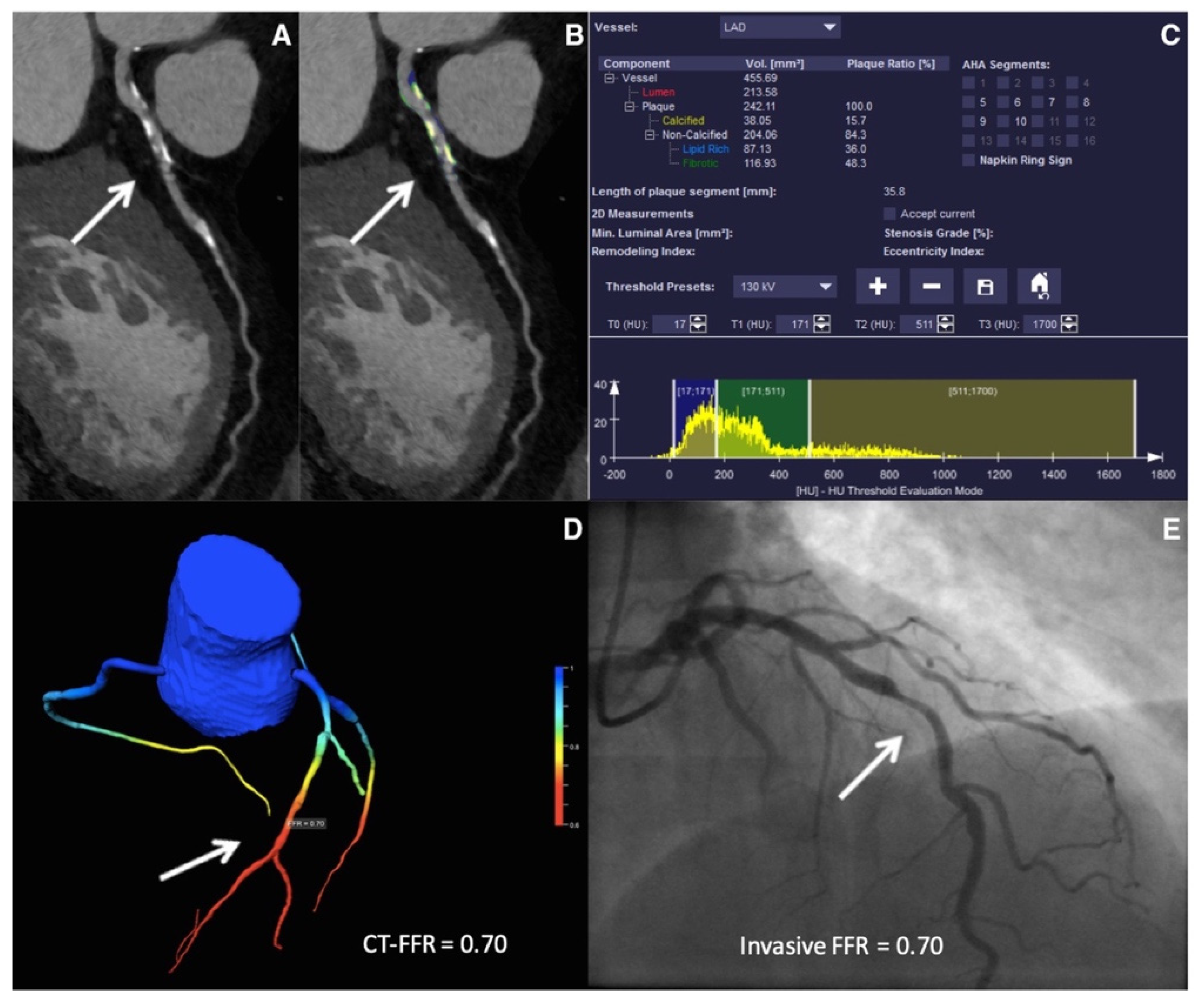
4. Artificial Intelligence to Predict Functionally Obstructive CAD and Lesion-Specific Ischemia—As a Gatekeeper to the Catheterization Laboratory
4.1. ML-Based CT-FFR Estimation and Diagnostic Accuracy
4.2. Impact of Calcification Burden on the Performance of CT-FFRML
4.3. CT-FFRML in Predicting Revascularization Events
5. Artificial Intelligence in the Field of Intracoronary Imaging
5.1. Artificial Intelligence to Optimize Peri-Intervention Workflow
5.2. Applications of Artificial Intelligence in Intra and Post-Intervention Workflow
6. Artificial Intelligence-Based Post-Procedure Risk Prediction Models
7. Artificial Intelligence-Based Long-Term Mortality and MACE Prediction Models
This entry is adapted from the peer-reviewed paper 10.3390/healthcare10020232
References
- Sprangers, M.A.G.; Sloan, J.A.; Barsevick, A.; Chauhan, C.; Dueck, A.C.; Raat, H.; Shi, Q.; Van Noorden, C.J.F.; Consortium, G. Scientific imperatives, clinical implications, and theoretical underpinnings for the investigation of the relationship between genetic variables and patient-reported quality-of-life outcomes. Qual. Life Res. 2010, 19, 1395–1403.
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403.
- Wang, Y.; Liu, T.; Liu, Y.; Chen, J.; Xin, B.; Wu, M.; Cui, W. Coronary artery disease associated specific modules and feature genes revealed by integrative methods of WGCNA, MetaDE and machine learning. Gene 2019, 710, 122–130.
- Balashanmugam, M.V.; Shivanandappa, T.B.; Nagarethinam, S.; Vastrad, B.; Vastrad, C. Analysis of Differentially Expressed Genes in Coronary Artery Disease by Integrated Microarray Analysis. Biomolecules 2019, 10, 35.
- Zhang, D.; Guan, L.; Li, X. Bioinformatics analysis identifies potential diagnostic signatures for coronary artery disease. J. Int. Med. Res. 2020, 48, 300060520979856.
- Ferguson, J.F.; Matthews, G.J.; Townsend, R.R.; Raj, D.S.; Kanetsky, P.A.; Budoff, M.; Fischer, M.J.; Rosas, S.E.; Kanthety, R.; Rahman, M.; et al. Candidate gene association study of coronary artery calcification in chronic kidney disease: Findings from the CRIC study (Chronic Renal Insufficiency Cohort). J. Am. Coll. Cardiol. 2013, 62, 789–798.
- O’Donnell, C.J.; Kavousi, M.; Smith, A.V.; Kardia, S.L.; Feitosa, M.F.; Hwang, S.J.; Sun, Y.V.; Province, M.A.; Aspelund, T.; Dehghan, A.; et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 2011, 124, 2855–2864.
- Diamond, G.A.; Forrester, J.S. Analysis of probability as an aid in the clinical diagnosis of coronary-artery disease. N. Engl. J. Med. 1979, 300, 1350–1358.
- Foldyna, B.; Udelson, J.E.; Karády, J.; Banerji, D.; Lu, M.T.; Mayrhofer, T.; Bittner, D.O.; Meyersohn, N.M.; Emami, H.; Genders, T.S.S.; et al. Pretest probability for patients with suspected obstructive coronary artery disease: Re-evaluating Diamond-Forrester for the contemporary era and clinical implications: Insights from the PROMISE trial. Eur. Heart J. Cardiovasc. Imaging 2019, 20, 574–581.
- Genders, T.S.; Steyerberg, E.W.; Alkadhi, H.; Leschka, S.; Desbiolles, L.; Nieman, K.; Galema, T.W.; Meijboom, W.B.; Mollet, N.R.; de Feyter, P.J.; et al. A clinical prediction rule for the diagnosis of coronary artery disease: Validation, updating, and extension. Eur. Heart J. 2011, 32, 1316–1330.
- Genders, T.S.; Steyerberg, E.W.; Hunink, M.G.; Nieman, K.; Galema, T.W.; Mollet, N.R.; de Feyter, P.J.; Krestin, G.P.; Alkadhi, H.; Leschka, S.; et al. Prediction model to estimate presence of coronary artery disease: Retrospective pooled analysis of existing cohorts. BMJ 2012, 344, e3485.
- Bittencourt, M.S.; Hulten, E.; Polonsky, T.S.; Hoffman, U.; Nasir, K.; Abbara, S.; Di Carli, M.; Blankstein, R. European Society of Cardiology-Recommended Coronary Artery Disease Consortium Pretest Probability Scores More Accurately Predict Obstructive Coronary Disease and Cardiovascular Events Than the Diamond and Forrester Score: The Partners Registry. Circulation 2016, 134, 201–211.
- Li, D.; Xiong, G.; Zeng, H.; Zhou, Q.; Jiang, J.; Guo, X. Machine learning-aided risk stratification system for the prediction of coronary artery disease. Int. J. Cardiol. 2021, 326, 30–34.
- Velusamy, D.; Ramasamy, K. Ensemble of heterogeneous classifiers for diagnosis and prediction of coronary artery disease with reduced feature subset. Comput. Methods Programs Biomed. 2021, 198, 105770.
- Muhammad, L.J.; Al-Shourbaji, I.; Haruna, A.A.; Mohammed, I.A.; Ahmad, A.; Jibrin, M.B. Machine Learning Predictive Models for Coronary Artery Disease. SN Comput. Sci. 2021, 2, 350.
- Lin, S.; Li, Z.; Fu, B.; Chen, S.; Li, X.; Wang, Y.; Wang, X.; Lv, B.; Xu, B.; Song, X.; et al. Feasibility of using deep learning to detect coronary artery disease based on facial photo. Eur. Heart J. 2020, 41, 4400–4411.
- Budoff, M.J.; Mayrhofer, T.; Ferencik, M.; Bittner, D.; Lee, K.L.; Lu, M.T.; Coles, A.; Jang, J.; Krishnam, M.; Douglas, P.S.; et al. Prognostic Value of Coronary Artery Calcium in the PROMISE Study (Prospective Multicenter Imaging Study for Evaluation of Chest Pain). Circulation 2017, 136, 1993–2005.
- Gulati, M.; Levy, P.D.; Mukherjee, D.; Amsterdam, E.; Bhatt, D.L.; Birtcher, K.K.; Blankstein, R.; Boyd, J.; Bullock-Palmer, R.P.; Conejo, T.; et al. 2021 AHA/ACC/ASE/CHEST/SAEM/SCCT/SCMR Guideline for the Evaluation and Diagnosis of Chest Pain: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2021, 78, e187–e285.
- Baskaran, L.; Ying, X.; Xu, Z.; Al’Aref, S.J.; Lee, B.C.; Lee, S.E.; Danad, I.; Park, H.B.; Bathina, R.; Baggiano, A.; et al. Machine learning insight into the role of imaging and clinical variables for the prediction of obstructive coronary artery disease and revascularization: An exploratory analysis of the CONSERVE study. PLoS ONE 2020, 15, e0233791.
- Al’Aref, S.J.; Maliakal, G.; Singh, G.; van Rosendael, A.R.; Ma, X.; Xu, Z.; Alawamlh, O.A.H.; Lee, B.; Pandey, M.; Achenbach, S.; et al. Machine learning of clinical variables and coronary artery calcium scoring for the prediction of obstructive coronary artery disease on coronary computed tomography angiography: Analysis from the CONFIRM registry. Eur. Heart J. 2020, 41, 359–367.
- Arsanjani, R.; Xu, Y.; Dey, D.; Fish, M.; Dorbala, S.; Hayes, S.; Berman, D.; Germano, G.; Slomka, P. Improved accuracy of myocardial perfusion SPECT for the detection of coronary artery disease using a support vector machine algorithm. J. Nucl. Med. 2013, 54, 549–555.
- Betancur, J.; Hu, L.H.; Commandeur, F.; Sharir, T.; Einstein, A.J.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.A.; Sinusas, A.J.; Miller, E.J.; et al. Deep Learning Analysis of Upright-Supine High-Efficiency SPECT Myocardial Perfusion Imaging for Prediction of Obstructive Coronary Artery Disease: A Multicenter Study. J. Nucl. Med. 2019, 60, 664–670.
- Guner, L.A.; Karabacak, N.I.; Akdemir, O.U.; Karagoz, P.S.; Kocaman, S.A.; Cengel, A.; Unlu, M. An open-source framework of neural networks for diagnosis of coronary artery disease from myocardial perfusion SPECT. J. Nucl. Cardiol. 2010, 17, 405–413.
- Rahmani, R.; Niazi, P.; Naseri, M.; Neishabouri, M.; Farzanefar, S.; Eftekhari, M.; Derakhshan, F.; Mollazadeh, R.; Meysami, A.; Abbasi, M. Improved diagnostic accuracy for myocardial perfusion imaging using artificial neural networks on different input variables including clinical and quantification data. Rev. Esp. Med. Nucl. E Imagen. Mol. 2019, 38, 275–279.
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J.; et al. Deep Learning for Prediction of Obstructive Disease From Fast Myocardial Perfusion SPECT: A Multicenter Study. JACC Cardiovasc. Imaging 2018, 11, 1654–1663.
- Arsanjani, R.; Xu, Y.; Dey, D.; Vahistha, V.; Shalev, A.; Nakanishi, R.; Hayes, S.; Fish, M.; Berman, D.; Germano, G.; et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J. Nucl. Cardiol. 2013, 20, 553–562.
- Rabbat, M.G.; Ramchandani, S.; Sanders, W.E., Jr. Cardiac Phase Space Analysis: Assessing Coronary Artery Disease Utilizing Artificial Intelligence. Biomed. Res. Int. 2021, 2021, 6637039.
- Stuckey, T.D.; Gammon, R.S.; Goswami, R.; Depta, J.P.; Steuter, J.A.; Meine, F.J., 3rd; Roberts, M.C.; Singh, N.; Ramchandani, S.; Burton, T.; et al. Cardiac Phase Space Tomography: A novel method of assessing coronary artery disease utilizing machine learning. PLoS ONE 2018, 13, e0198603.
- Medina, R.; Panidis, I.P.; Morganroth, J.; Kotler, M.N.; Mintz, G.S. The value of echocardiographic regional wall motion abnormalities in detecting coronary artery disease in patients with or without a dilated left ventricle. Am. Heart J. 1985, 109, 799–803.
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Oh, S.L.; Adam, M.; Koh, J.E.W.; Tan, J.H.; Ghista, D.N.; Martis, R.J.; Chua, C.K.; et al. Automated detection and localization of myocardial infarction using electrocardiogram: A comparative study of different leads. Knowl.-Based Syst. 2016, 99, 146–156.
- Han, C.; Shi, L. ML–ResNet: A novel network to detect and locate myocardial infarction using 12 leads ECG. Comput. Methods Programs Biomed. 2020, 185, 105138.
- Lih, O.S.; Jahmunah, V.; San, T.R.; Ciaccio, E.J.; Yamakawa, T.; Tanabe, M.; Kobayashi, M.; Faust, O.; Acharya, U.R. Comprehensive electrocardiographic diagnosis based on deep learning. Artif. Intell. Med. 2020, 103, 101789.
- Liu, C.Y.; Tang, C.X.; Zhang, X.L.; Chen, S.; Xie, Y.; Zhang, X.Y.; Qiao, H.Y.; Zhou, C.S.; Xu, P.P.; Lu, M.J.; et al. Deep learning powered coronary CT angiography for detecting obstructive coronary artery disease: The effect of reader experience, calcification and image quality. Eur. J. Radiol. 2021, 142, 109835.
- Lee, J.-G.; Kim, H.; Kang, H.; Koo, H.J.; Kang, J.-W.; Kim, Y.-H.; Yang, D.H. Fully Automatic Coronary Calcium Score Software Empowered by Artificial Intelligence Technology: Validation Study Using Three CT Cohorts. Korean J. Radiol. 2021, 22, 1764–1776.
- van Velzen, S.G.M.; Lessmann, N.; Velthuis, B.K.; Bank, I.E.M.; van den Bongard, D.; Leiner, T.; de Jong, P.A.; Veldhuis, W.B.; Correa, A.; Terry, J.G.; et al. Deep Learning for Automatic Calcium Scoring in CT: Validation Using Multiple Cardiac CT and Chest CT Protocols. Radiology 2020, 295, 66–79.
- Baskaran, L.; Maliakal, G.; Al’Aref, S.J.; Singh, G.; Xu, Z.; Michalak, K.; Dolan, K.; Gianni, U.; van Rosendael, A.; van den Hoogen, I.; et al. Identification and Quantification of Cardiovascular Structures From CCTA: An End-to-End, Rapid, Pixel-Wise, Deep-Learning Method. JACC Cardiovasc. Imaging 2020, 13, 1163–1171.
- Wang, W.; Wang, H.; Chen, Q.; Zhou, Z.; Wang, R.; Wang, H.; Zhang, N.; Chen, Y.; Sun, Z.; Xu, L. Coronary artery calcium score quantification using a deep-learning algorithm. Clin. Radiol. 2020, 75, 237.e11–237.e16.
- von Knebel Doeberitz, P.L.; De Cecco, C.N.; Schoepf, U.J.; Duguay, T.M.; Albrecht, M.H.; van Assen, M.; Bauer, M.J.; Savage, R.H.; Pannell, J.T.; De Santis, D.; et al. Coronary CT angiography-derived plaque quantification with artificial intelligence CT fractional flow reserve for the identification of lesion-specific ischemia. Eur. Radiol. 2019, 29, 2378–2387.
- Koo, H.J.; Lee, J.G.; Ko, J.Y.; Lee, G.; Kang, J.W.; Kim, Y.H.; Yang, D.H. Automated Segmentation of Left Ventricular Myocardium on Cardiac Computed Tomography Using Deep Learning. Korean J. Radiol. 2020, 21, 660–669.
- Morris, E.D.; Ghanem, A.I.; Dong, M.; Pantelic, M.V.; Walker, E.M.; Glide-Hurst, C.K. Cardiac substructure segmentation with deep learning for improved cardiac sparing. Med. Phys. 2020, 47, 576–586.
- Muscogiuri, G.; Chiesa, M.; Trotta, M.; Gatti, M.; Palmisano, V.; Dell’Aversana, S.; Baessato, F.; Cavaliere, A.; Cicala, G.; Loffreno, A.; et al. Performance of a deep learning algorithm for the evaluation of CAD-RADS classification with CCTA. Atherosclerosis 2020, 294, 25–32.
- Fihn, S.D.; Gardin, J.M.; Abrams, J.; Berra, K.; Blankenship, J.C.; Dallas, A.P.; Douglas, P.S.; Foody, J.M.; Gerber, T.C.; Hinderliter, A.L.; et al. 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease. Circulation 2012, 126, e354–e471.
- Biagini, E.; Shaw, L.J.; Poldermans, D.; Schinkel, A.F.; Rizzello, V.; Elhendy, A.; Rapezzi, C.; Bax, J.J. Accuracy of non-invasive techniques for diagnosis of coronary artery disease and prediction of cardiac events in patients with left bundle branch block: A meta-analysis. Eur. J. Nucl. Med. Mol. Imaging 2006, 33, 1442–1451.
- Mahajan, N.; Polavaram, L.; Vankayala, H.; Ference, B.; Wang, Y.; Ager, J.; Kovach, J.; Afonso, L. Diagnostic accuracy of myocardial perfusion imaging and stress echocardiography for the diagnosis of left main and triple vessel coronary artery disease: A comparative meta-analysis. Heart 2010, 96, 956–966.
- Jaarsma, C.; Leiner, T.; Bekkers Sebastiaan, C.; Crijns Harry, J.; Wildberger Joachim, E.; Nagel, E.; Nelemans Patricia, J.; Schalla, S. Diagnostic Performance of Noninvasive Myocardial Perfusion Imaging Using Single-Photon Emission Computed Tomography, Cardiac Magnetic Resonance, and Positron Emission Tomography Imaging for the Detection of Obstructive Coronary Artery Disease. J. Am. Coll. Cardiol. 2012, 59, 1719–1728.
- Takx, R.A.P.; Blomberg, B.A.; Aidi, H.E.; Habets, J.; de Jong, P.A.; Nagel, E.; Hoffmann, U.; Leiner, T. Diagnostic Accuracy of Stress Myocardial Perfusion Imaging Compared to Invasive Coronary Angiography With Fractional Flow Reserve Meta-Analysis. Circ. Cardiovasc. Imaging 2015, 8, e002666.
- Fleischmann, K.E.; Hunink, M.G.; Kuntz, K.M.; Douglas, P.S. Exercise echocardiography or exercise SPECT imaging? A meta-analysis of diagnostic test performance. JAMA 1998, 280, 913–920.
- Holder, L.; Lewis, S.; Abrames, E.; Wolin, E.A. Review of SPECT myocardial perfusion imaging. J. Am. Osteopath. Coll. Radiol. 2016, 5, 5–13.
- Czaja, M.; Wygoda, Z.; Duszańska, A.; Szczerba, D.; Głowacki, J.; Gąsior, M.; Wasilewski, J.P. Interpreting myocardial perfusion scintigraphy using single-photon emission computed tomography. Part 1. Kardiochir. Torakochirurgia Pol. 2017, 14, 192–199.
- Slomka, P.; Xu, Y.; Berman, D.; Germano, G. Quantitative analysis of perfusion studies: Strengths and pitfalls. J. Nucl. Cardiol. Off. Publ. Am. Soc. Nucl. Cardiol. 2012, 19, 338–346.
- Hachamovitch, R.; Hayes, S.W.; Friedman, J.D.; Cohen, I.; Berman, D.S. A prognostic score for prediction of cardiac mortality risk after adenosine stress myocardial perfusion scintigraphy. J. Am. Coll. Cardiol. 2005, 45, 722–729.
- Arsanjani, R.; Xu, Y.; Hayes, S.W.; Fish, M.; Lemley, M., Jr.; Gerlach, J.; Dorbala, S.; Berman, D.S.; Germano, G.; Slomka, P. Comparison of fully automated computer analysis and visual scoring for detection of coronary artery disease from myocardial perfusion SPECT in a large population. J. Nucl. Med. 2013, 54, 221–228.
- Hu, L.H.; Betancur, J.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.A.; Sinusas, A.J.; Miller, E.J.; et al. Machine learning predicts per-vessel early coronary revascularization after fast myocardial perfusion SPECT: Results from multicentre REFINE SPECT registry. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 549–559.
- Arsanjani, R.; Dey, D.; Khachatryan, T.; Shalev, A.; Hayes, S.W.; Fish, M.; Nakanishi, R.; Germano, G.; Berman, D.S.; Slomka, P. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J. Nucl. Cardiol. 2015, 22, 877–884.
- Itu, L.; Rapaka, S.; Passerini, T.; Georgescu, B.; Schwemmer, C.; Schoebinger, M.; Flohr, T.; Sharma, P.; Comaniciu, D. A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. J. Appl. Physiol. 2016, 121, 42–52.
- Coenen, A.; Kim, Y.H.; Kruk, M.; Tesche, C.; De Geer, J.; Kurata, A.; Lubbers, M.L.; Daemen, J.; Itu, L.; Rapaka, S.; et al. Diagnostic Accuracy of a Machine-Learning Approach to Coronary Computed Tomographic Angiography-Based Fractional Flow Reserve: Result From the MACHINE Consortium. Circ. Cardiovasc. Imaging 2018, 11, e007217.
- Di Jiang, M.; Zhang, X.L.; Liu, H.; Tang, C.X.; Li, J.H.; Wang, Y.N.; Xu, P.P.; Zhou, C.S.; Zhou, F.; Lu, M.J.; et al. The effect of coronary calcification on diagnostic performance of machine learning-based CT-FFR: A Chinese multicenter study. Eur. Radiol. 2021, 31, 1482–1493.
- Koo, H.J.; Kang, J.W.; Kang, S.J.; Kweon, J.; Lee, J.G.; Ahn, J.M.; Park, D.W.; Lee, S.W.; Lee, C.W.; Park, S.W.; et al. Impact of coronary calcium score and lesion characteristics on the diagnostic performance of machine-learning-based computed tomography-derived fractional flow reserve. Eur. Heart J. Cardiovasc. Imaging 2021, 22, 998–1006.
- Kumamaru, K.K.; Fujimoto, S.; Otsuka, Y.; Kawasaki, T.; Kawaguchi, Y.; Kato, E.; Takamura, K.; Aoshima, C.; Kamo, Y.; Kogure, Y.; et al. Diagnostic accuracy of 3D deep-learning-based fully automated estimation of patient-level minimum fractional flow reserve from coronary computed tomography angiography. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 437–445.
- Kurata, A.; Fukuyama, N.; Hirai, K.; Kawaguchi, N.; Tanabe, Y.; Okayama, H.; Shigemi, S.; Watanabe, K.; Uetani, T.; Ikeda, S.; et al. On-Site Computed Tomography-Derived Fractional Flow Reserve Using a Machine-Learning Algorithm—Clinical Effectiveness in a Retrospective Multicenter Cohort. Circ. J. 2019, 83, 1563–1571.
- Rother, J.; Moshage, M.; Dey, D.; Schwemmer, C.; Trobs, M.; Blachutzik, F.; Achenbach, S.; Schlundt, C.; Marwan, M. Comparison of invasively measured FFR with FFR derived from coronary CT angiography for detection of lesion-specific ischemia: Results from a PC-based prototype algorithm. J. Cardiovasc. Comput. Tomogr. 2018, 12, 101–107.
- Tang, C.X.; Wang, Y.N.; Zhou, F.; Schoepf, U.J.; Assen, M.V.; Stroud, R.E.; Li, J.H.; Zhang, X.L.; Lu, M.J.; Zhou, C.S.; et al. Diagnostic performance of fractional flow reserve derived from coronary CT angiography for detection of lesion-specific ischemia: A multi-center study and meta-analysis. Eur. J. Radiol. 2019, 116, 90–97.
- Tesche, C.; Otani, K.; De Cecco, C.N.; Coenen, A.; De Geer, J.; Kruk, M.; Kim, Y.H.; Albrecht, M.H.; Baumann, S.; Renker, M.; et al. Influence of Coronary Calcium on Diagnostic Performance of Machine Learning CT-FFR: Results From MACHINE Registry. JACC Cardiovasc. Imaging 2020, 13, 760–770.
- Wang, Z.Q.; Zhou, Y.J.; Zhao, Y.X.; Shi, D.M.; Liu, Y.Y.; Liu, W.; Liu, X.L.; Li, Y.P. Diagnostic accuracy of a deep learning approach to calculate FFR from coronary CT angiography. J. Geriatr. Cardiol. 2019, 16, 42–48.
- Wardziak, L.; Kruk, M.; Pleban, W.; Demkow, M.; Ruzyllo, W.; Dzielinska, Z.; Kepka, C. Coronary CTA enhanced with CTA based FFR analysis provides higher diagnostic value than invasive coronary angiography in patients with intermediate coronary stenosis. J. Cardiovasc. Comput. Tomogr. 2019, 13, 62–67.
- Tesche, C.; De Cecco, C.N.; Baumann, S.; Renker, M.; McLaurin, T.W.; Duguay, T.M.; Bayer, R.R., 2nd; Steinberg, D.H.; Grant, K.L.; Canstein, C.; et al. Coronary CT Angiography-derived Fractional Flow Reserve: Machine Learning Algorithm versus Computational Fluid Dynamics Modeling. Radiology 2018, 288, 64–72.
- Arbab-Zadeh, A.; Miller, J.M.; Rochitte, C.E.; Dewey, M.; Niinuma, H.; Gottlieb, I.; Paul, N.; Clouse, M.E.; Shapiro, E.P.; Hoe, J.; et al. Diagnostic accuracy of computed tomography coronary angiography according to pre-test probability of coronary artery disease and severity of coronary arterial calcification. The CORE-64 (Coronary Artery Evaluation Using 64-Row Multidetector Computed Tomography Angiography) International Multicenter Study. J. Am. Coll. Cardiol. 2012, 59, 379–387.
- Chen, C.-C.; Chen, C.-C.; Hsieh, I.C.; Liu, Y.-C.; Liu, C.-Y.; Chan, T.; Wen, M.-S.; Wan, Y.-L. The effect of calcium score on the diagnostic accuracy of coronary computed tomography angiography. Int. J. Cardiovasc. Imaging 2011, 27, 37–42.
- Vavere, A.L.; Arbab-Zadeh, A.; Rochitte, C.E.; Dewey, M.; Niinuma, H.; Gottlieb, I.; Clouse, M.E.; Bush, D.E.; Hoe, J.W.M.; de Roos, A.; et al. Coronary artery stenoses: Accuracy of 64-detector row CT angiography in segments with mild, moderate, or severe calcification--a subanalysis of the CORE-64 trial. Radiology 2011, 261, 100–108.
- Arjmand Shabestari, A. Coronary artery calcium score: A review. Iran Red. Crescent. Med. J. 2013, 15, e16616.
- Agatston, A.S.; Janowitz, W.R.; Hildner, F.J.; Zusmer, N.R.; Viamonte, M., Jr.; Detrano, R. Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 1990, 15, 827–832.
- Yu, M.; Li, Y.; Li, W.; Lu, Z.; Wei, M.; Zhang, J. Calcification remodeling index assessed by cardiac CT predicts severe coronary stenosis in lesions with moderate to severe calcification. J. Cardiovasc. Comput. Tomogr. 2018, 12, 42–49.
- Sekimoto, T.; Akutsu, Y.; Hamazaki, Y.; Sakai, K.; Kosaki, R.; Yokota, H.; Tsujita, H.; Tsukamoto, S.; Kaneko, K.; Sakurai, M.; et al. Regional calcified plaque score evaluated by multidetector computed tomography for predicting the addition of rotational atherectomy during percutaneous coronary intervention. J. Cardiovasc. Comput. Tomogr. 2016, 10, 221–228.
- Qiao, H.Y.; Tang, C.X.; Schoepf, U.J.; Tesche, C.; Bayer, R.R., 2nd; Giovagnoli, D.A.; Todd Hudson, H., Jr.; Zhou, C.S.; Yan, J.; Lu, M.J.; et al. Impact of machine learning-based coronary computed tomography angiography fractional flow reserve on treatment decisions and clinical outcomes in patients with suspected coronary artery disease. Eur. Radiol. 2020, 30, 5841–5851.
- Liu, X.; Mo, X.; Zhang, H.; Yang, G.; Shi, C.; Hau, W.K. A 2-year investigation of the impact of the computed tomography-derived fractional flow reserve calculated using a deep learning algorithm on routine decision-making for coronary artery disease management. Eur. Radiol. 2021, 31, 7039–7046.
- Martin, S.S.; Mastrodicasa, D.; van Assen, M.; De Cecco, C.N.; Bayer, R.R.; Tesche, C.; Varga-Szemes, A.; Fischer, A.M.; Jacobs, B.E.; Sahbaee, P.; et al. Value of Machine Learning-based Coronary CT Fractional Flow Reserve Applied to Triple-Rule-Out CT Angiography in Acute Chest Pain. Radiol. Cardiothorac. Imaging 2020, 2, e190137.
- Nous, F.M.A.; Budde, R.P.J.; Lubbers, M.M.; Yamasaki, Y.; Kardys, I.; Bruning, T.A.; Akkerhuis, J.M.; Kofflard, M.J.M.; Kietselaer, B.; Galema, T.W.; et al. Impact of machine-learning CT-derived fractional flow reserve for the diagnosis and management of coronary artery disease in the randomized CRESCENT trials. Eur. Radiol. 2020, 30, 3692–3701.
- Cook, C.M.; Petraco, R.; Shun-Shin, M.J.; Ahmad, Y.; Nijjer, S.; Al-Lamee, R.; Kikuta, Y.; Shiono, Y.; Mayet, J.; Francis, D.P.; et al. Diagnostic Accuracy of Computed Tomography–Derived Fractional Flow Reserve: A Systematic Review. JAMA Cardiol. 2017, 2, 803–810.
- Gaur, S.; Ovrehus, K.A.; Dey, D.; Leipsic, J.; Botker, H.E.; Jensen, J.M.; Narula, J.; Ahmadi, A.; Achenbach, S.; Ko, B.S.; et al. Coronary plaque quantification and fractional flow reserve by coronary computed tomography angiography identify ischaemia-causing lesions. Eur. Heart J. 2016, 37, 1220–1227.
- Kawasaki, T.; Kidoh, M.; Kido, T.; Sueta, D.; Fujimoto, S.; Kumamaru, K.K.; Uetani, T.; Tanabe, Y.; Ueda, T.; Sakabe, D.; et al. Evaluation of Significant Coronary Artery Disease Based on CT Fractional Flow Reserve and Plaque Characteristics Using Random Forest Analysis in Machine Learning. Acad. Radiol. 2020, 27, 1700–1708.
- Bae, Y.; Kang, S.J.; Kim, G.; Lee, J.G.; Min, H.S.; Cho, H.; Kang, D.Y.; Lee, P.H.; Ahn, J.M.; Park, D.W.; et al. Prediction of coronary thin-cap fibroatheroma by intravascular ultrasound-based machine learning. Atherosclerosis 2019, 288, 168–174.
- Min, H.S.; Yoo, J.H.; Kang, S.J.; Lee, J.G.; Cho, H.; Lee, P.H.; Ahn, J.M.; Park, D.W.; Lee, S.W.; Kim, Y.H.; et al. Detection of optical coherence tomography-defined thin-cap fibroatheroma in the coronary artery using deep learning. EuroIntervention 2020, 16, 404–412.
- Maehara, A.; Matsumura, M.; Ali, Z.A.; Mintz, G.S.; Stone, G.W. IVUS-Guided Versus OCT-Guided Coronary Stent Implantation: A Critical Appraisal. JACC Cardiovasc. Imaging 2017, 10, 1487–1503.
- Nishi, T.; Yamashita, R.; Imura, S.; Tateishi, K.; Kitahara, H.; Kobayashi, Y.; Yock, P.G.; Fitzgerald, P.J.; Honda, Y. Deep learning-based intravascular ultrasound segmentation for the assessment of coronary artery disease. Int. J. Cardiol. 2021, 333, 55–59.
- Brown, A.J.; Teng, Z.; Calvert, P.A.; Rajani, N.K.; Hennessy, O.; Nerlekar, N.; Obaid, D.R.; Costopoulos, C.; Huang, Y.; Hoole, S.P.; et al. Plaque Structural Stress Estimations Improve Prediction of Future Major Adverse Cardiovascular Events After Intracoronary Imaging. Circ. Cardiovasc. Imaging 2016, 9, e004172.
- Xie, Z.; Dong, N.; Sun, R.; Liu, X.; Gu, X.; Sun, Y.; Du, H.; Dai, J.; Liu, Y.; Hou, J.; et al. Relation between baseline plaque features and subsequent coronary artery remodeling determined by optical coherence tomography and intravascular ultrasound. Oncotarget 2017, 8, 4234–4244.
- Stone, P.H.; Saito, S.; Takahashi, S.; Makita, Y.; Nakamura, S.; Kawasaki, T.; Takahashi, A.; Katsuki, T.; Nakamura, S.; Namiki, A.; et al. Prediction of progression of coronary artery disease and clinical outcomes using vascular profiling of endothelial shear stress and arterial plaque characteristics: The PREDICTION Study. Circulation 2012, 126, 172–181.
- Calvert, P.A.; Obaid, D.R.; O’Sullivan, M.; Shapiro, L.M.; McNab, D.; Densem, C.G.; Schofield, P.M.; Braganza, D.; Clarke, S.C.; Ray, K.K.; et al. Association between IVUS findings and adverse outcomes in patients with coronary artery disease: The VIVA (VH-IVUS in Vulnerable Atherosclerosis) Study. JACC Cardiovasc. Imaging 2011, 4, 894–901.
- Zhang, L.; Wahle, A.; Chen, Z.; Lopez, J.J.; Kovarnik, T.; Sonka, M. Predicting Locations of High-Risk Plaques in Coronary Arteries in Patients Receiving Statin Therapy. IEEE Trans. Med. Imaging 2018, 37, 151–161.
- Farooq, V.; Brugaletta, S.; Serruys, P.W. The SYNTAX score and SYNTAX-based clinical risk scores. Semin Thorac Cardiovasc Surg 2011, 23, 99–105.
- Singh, M.; Rihal, C.S.; Lennon, R.J.; Spertus, J.; Rumsfeld, J.S.; Holmes, D.R., Jr. Bedside estimation of risk from percutaneous coronary intervention: The new Mayo Clinic risk scores. Mayo Clin. Proc. 2007, 82, 701–708.
- Chowdhary, S.; Ivanov, J.; Mackie, K.; Seidelin, P.H.; Dzavík, V. The Toronto score for in-hospital mortality after percutaneous coronary interventions. Am. Heart J. 2009, 157, 156–163.
- Hannan, E.L.; Farrell, L.S.; Walford, G.; Jacobs, A.K.; Berger, P.B.; Holmes, D.R., Jr.; Stamato, N.J.; Sharma, S.; King, S.B., 3rd. The New York State risk score for predicting in-hospital/30-day mortality following percutaneous coronary intervention. JACC Cardiovasc. Interv. 2013, 6, 614–622.
- MacKenzie, T.A.; Malenka, D.J.; Olmstead, E.M.; Piper, W.D.; Langner, C.; Ross, C.S.; O’Connor, G.T. Prediction of survival after coronary revascularization: Modeling short-term, mid-term, and long-term survival. Ann. Thorac. Surg. 2009, 87, 463–472.
- O’Connor, G.T.; Malenka, D.J.; Quinton, H.; Robb, J.F.; Kellett, M.A., Jr.; Shubrooks, S.; Bradley, W.A.; Hearne, M.J.; Watkins, M.W.; Wennberg, D.E.; et al. Multivariate prediction of in-hospital mortality after percutaneous coronary interventions in 1994-1996. Northern New England Cardiovascular Disease Study Group. J. Am. Coll. Cardiol. 1999, 34, 681–691.
- Rihal, C.S.; Grill, D.E.; Bell, M.R.; Berger, P.B.; Garratt, K.N.; Holmes, D.R., Jr. Prediction of death after percutaneous coronary interventional procedures. Am. Heart J. 2000, 139, 1032–1038.
- Wu, C.; Hannan, E.L.; Walford, G.; Ambrose, J.A.; Holmes, D.R., Jr.; King, S.B., 3rd; Clark, L.T.; Katz, S.; Sharma, S.; Jones, R.H. A risk score to predict in-hospital mortality for percutaneous coronary interventions. J. Am. Coll. Cardiol. 2006, 47, 654–660.
- Fanaroff, A.C.; Zakroysky, P.; Dai, D.; Wojdyla, D.; Sherwood, M.W.; Roe, M.T.; Wang, T.Y.; Peterson, E.D.; Gurm, H.S.; Cohen, M.G.; et al. Outcomes of PCI in Relation to Procedural Characteristics and Operator Volumes in the United States. J. Am. Coll. Cardiol. 2017, 69, 2913–2924.
- Iverson, A.; Stanberry, L.I.; Tajti, P.; Garberich, R.; Antos, A.; Burke, M.N.; Chavez, I.; Gössl, M.; Henry, T.D.; Lips, D.; et al. Prevalence, Trends, and Outcomes of Higher-Risk Percutaneous Coronary Interventions Among Patients without Acute Coronary Syndromes. Cardiovasc. Revasc. Med. 2019, 20, 289–292.
- Singh, M.; Lennon, R.J.; Gulati, R.; Holmes, D.R. Risk scores for 30-day mortality after percutaneous coronary intervention: New insights into causes and risk of death. Mayo Clin. Proc. 2014, 89, 631–637.
- Steele, A.J.; Denaxas, S.C.; Shah, A.D.; Hemingway, H.; Luscombe, N.M. Machine learning models in electronic health records can outperform conventional survival models for predicting patient mortality in coronary artery disease. PLoS ONE 2018, 13, e0202344.
- Bertsimas, D.; Orfanoudaki, A.; Weiner, R.B. Personalized treatment for coronary artery disease patients: A machine learning approach. Health Care Manag. Sci. 2020, 23, 482–506.
- Farhadian, M.; Dehdar Karsidani, S.; Mozayanimonfared, A.; Mahjub, H. Risk factors associated with major adverse cardiac and cerebrovascular events following percutaneous coronary intervention: A 10-year follow-up comparing random survival forest and Cox proportional-hazards model. BMC Cardiovasc. Disord. 2021, 21, 38.
- Krittanawong, C.; Zhang, H.; Wang, Z.; Aydar, M.; Kitai, T. Artificial Intelligence in Precision Cardiovascular Medicine. J. Am. Coll. Cardiol. 2017, 69, 2657–2664.
- Chao, H.; Shan, H.; Homayounieh, F.; Singh, R.; Khera, R.D.; Guo, H.; Su, T.; Wang, G.; Kalra, M.K.; Yan, P. Deep learning predicts cardiovascular disease risks from lung cancer screening low dose computed tomography. Nat. Commun. 2021, 12, 2963.
- Wolterink, J.; Leiner, T.; Takx, R.A.; Viergever, M.; Išgum, I. An Automatic Machine Learning System for Coronary Calcium Scoring in Clinical Non-Contrast Enhanced, ECG-Triggered Cardiac CT; SPIE: San Diego, CA, USA, 2014; Volume 9035.
- Sandstedt, M.; Henriksson, L.; Janzon, M.; Nyberg, G.; Engvall, J.; De Geer, J.; Alfredsson, J.; Persson, A. Evaluation of an AI-based, automatic coronary artery calcium scoring software. Eur. Radiol. 2020, 30, 1671–1678.
- Nakanishi, R.; Rajani, R.; Cheng, V.Y.; Gransar, H.; Nakazato, R.; Shmilovich, H.; Otaki, Y.; Hayes, S.W.; Thomson, L.E.; Friedman, J.D.; et al. Increase in epicardial fat volume is associated with greater coronary artery calcification progression in subjects at intermediate risk by coronary calcium score: A serial study using non-contrast cardiac CT. Atherosclerosis 2011, 218, 363–368.
- Commandeur, F.; Slomka, P.J.; Goeller, M.; Chen, X.; Cadet, S.; Razipour, A.; McElhinney, P.; Gransar, H.; Cantu, S.; Miller, R.J.H.; et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: A prospective study. Cardiovasc. Res. 2020, 116, 2216–2225.
- Eisenberg, E.; McElhinney, P.A.; Commandeur, F.; Chen, X.; Cadet, S.; Goeller, M.; Razipour, A.; Gransar, H.; Cantu, S.; Miller, R.J.H.; et al. Deep Learning-Based Quantification of Epicardial Adipose Tissue Volume and Attenuation Predicts Major Adverse Cardiovascular Events in Asymptomatic Subjects. Circ. Cardiovasc. Imaging 2020, 13, e009829.
- Han, D.; Kolli, K.K.; Gransar, H.; Lee, J.H.; Choi, S.Y.; Chun, E.J.; Han, H.W.; Park, S.H.; Sung, J.; Jung, H.O.; et al. Machine learning based risk prediction model for asymptomatic individuals who underwent coronary artery calcium score: Comparison with traditional risk prediction approaches. J. Cardiovasc. Comput. Tomogr. 2020, 14, 168–176.
- Tamarappoo, B.K.; Lin, A.; Commandeur, F.; McElhinney, P.A.; Cadet, S.; Goeller, M.; Razipour, A.; Chen, X.; Gransar, H.; Cantu, S.; et al. Machine learning integration of circulating and imaging biomarkers for explainable patient-specific prediction of cardiac events: A prospective study. Atherosclerosis 2021, 318, 76–82.
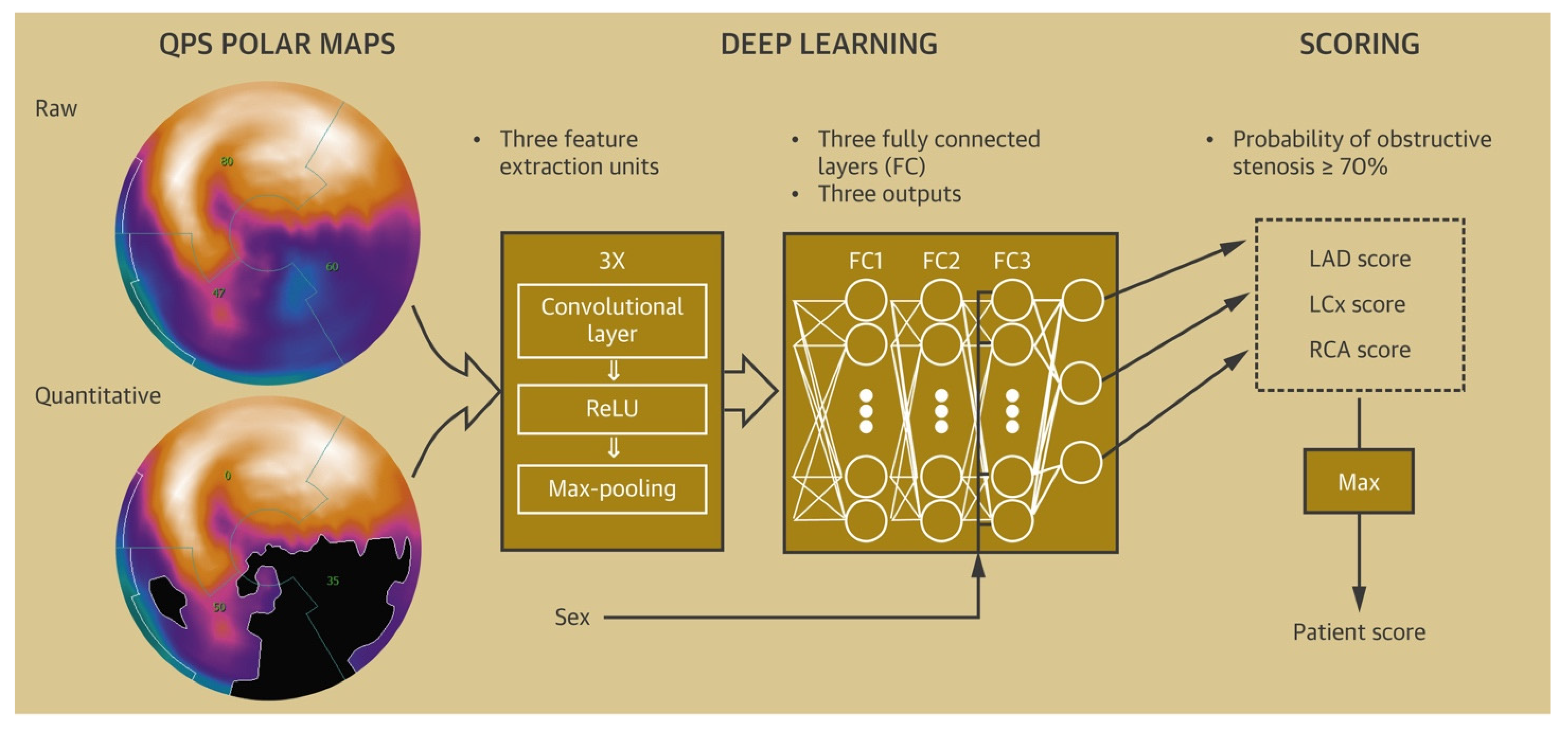
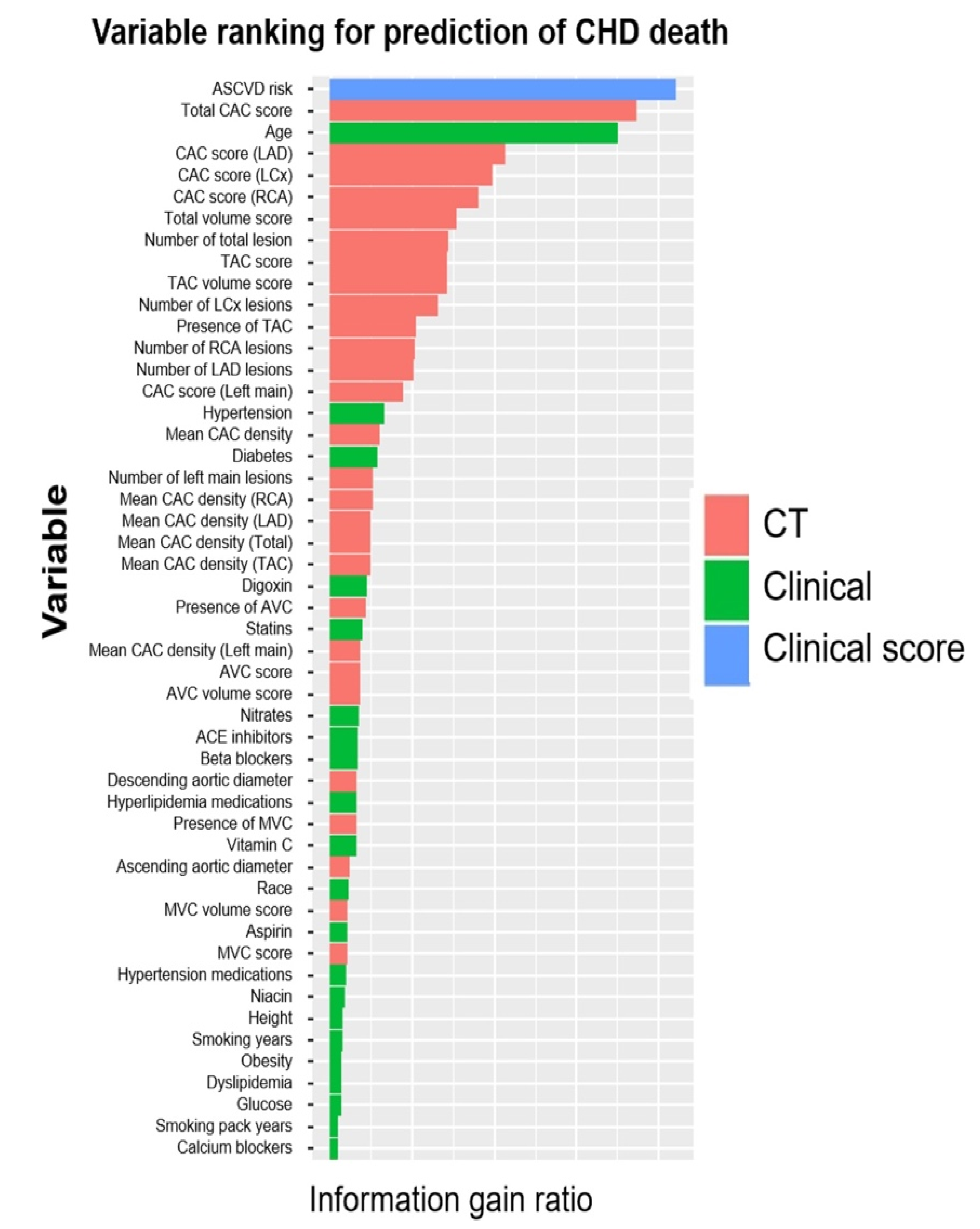
- Nakanishi, R.; Slomka, P.J.; Rios, R.; Betancur, J.; Blaha, M.J.; Nasir, K.; Miedema, M.D.; Rumberger, J.A.; Gransar, H.; Shaw, L.J.; et al. Machine Learning Adds to Clinical and CAC Assessments in Predicting 10-Year CHD and CVD Deaths. JACC Cardiovasc. Imaging 2021, 14, 615–625.
- Antonopoulos, A.S.; Sanna, F.; Sabharwal, N.; Thomas, S.; Oikonomou, E.K.; Herdman, L.; Margaritis, M.; Shirodaria, C.; Kampoli, A.M.; Akoumianakis, I.; et al. Detecting human coronary inflammation by imaging perivascular fat. Sci. Transl. Med. 2017, 9, eaal2658.
- Oikonomou, E.K.; Marwan, M.; Desai, M.Y.; Mancio, J.; Alashi, A.; Hutt Centeno, E.; Thomas, S.; Herdman, L.; Kotanidis, C.P.; Thomas, K.E.; et al. Non-invasive detection of coronary inflammation using computed tomography and prediction of residual cardiovascular risk (the CRISP CT study): A post-hoc analysis of prospective outcome data. Lancet 2018, 392, 929–939.
- Oikonomou Evangelos, K.; Desai Milind, Y.; Marwan, M.; Kotanidis Christos, P.; Antonopoulos Alexios, S.; Schottlander, D.; Channon Keith, M.; Neubauer, S.; Achenbach, S.; Antoniades, C. Perivascular Fat Attenuation Index Stratifies Cardiac Risk Associated with High-Risk Plaques in the CRISP-CT Study. J. Am. Coll. Cardiol. 2020, 76, 755–757.
- Oikonomou, E.K.; Williams, M.C.; Kotanidis, C.P.; Desai, M.Y.; Marwan, M.; Antonopoulos, A.S.; Thomas, K.E.; Thomas, S.; Akoumianakis, I.; Fan, L.M.; et al. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur. Heart J. 2019, 40, 3529–3543.
