Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Oncology
Due to the need for quality biomarkers for triple-negative breast cancer (TNBC) because of its aggressive nature and limited therapeutic options, it is not surprising that several studies have focused on identifying expression differences in peripheral blood cells between TNBC patients and other classical breast cancer (BC) subtypes.
- triple negative breast cancer
1. Gene Expression Biomarkers in Nucleated Blood Cells of TNBC Patients
Due to the need for quality biomarkers for TNBC because of its aggressive nature and limited therapeutic options, it is not surprising that several studies have focused on identifying expression differences in peripheral blood cells between TNBC patients and other classical BC subtypes. Despite the described heterogeneity of PBMC transcriptomes in breast cancer, some characteristic molecular features for the TNBC subtype have been reported (Figure 3 and Table 2), as detailed below. The main common observation was that an extensive immune response and tumor-related inflammation are strongly involved in TNBC.
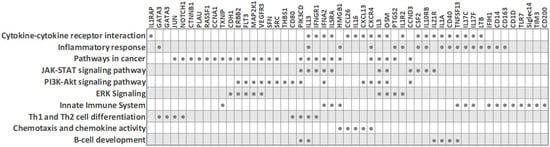
Figure 3. Biological processes and transcripts significantly deregulated in peripheral blood cells of TNBC patients compared to other BC subtypes (based on PBC transcriptome studies). The filled circles mark the transcripts that are differentially expressed in TNBC.
Table 2. Peripheral blood transcriptome studies describing gene expression signatures in TNBC patients.
| Study Cohort | Methodology | Gene Signatures in TNBC Patients | Reference |
|---|---|---|---|
| 13 BC (4 ER+/HER2-, 2ER+/HER2+, 3 ER-/HER2+, 4 ER-/HER2-) and 3 healthy subjects |
RNA-seq | Distinct transcriptome in 3 ER-HER2- patients (i.e., monocyte activating immune subgroup): upregulated CD14, CD40 (TNFRSF5), CD80, Siglec14, NRP1, TIM3 |
[89] |
| 29 BC (14 TNBC and 15 hormone-dependent (ER+/PR+/HER2-)) and 7 healthy subjects |
Microarray analysis | Thirty-four-gene TNBC signature (distinguishing TNBC from both ER+/PR+/HER2- and healthy controls); downregulated genes: RNU105A, SCARNA5, CD200, SNORA53, BICC1, KLHL31, SNORA81, FAM86DP, SNORD3B-1, RNU2-2, LMAN1, STXBP4, MGC57346, MAP7D2, CCDC39, SNORD15A, SNORD3B-1, ZNF3, SNORD17, SNORA12, NT5E, SNORA74A, NT5E, SCARNA6; upregulated genes: PLAU, LOC100128175, ITPK1-AS1, ALPK3, C10orf105, ASAP1-IT1 |
[5] |
| 23 BC (14 TNBC and 9 luminal-A) |
Pan-Cancer Immune Profiling Panel, 770 genes | Distinct transcriptome in 3 out of 14 TNBC patients; upregulated genes: IL1R2, THBS1, CD163, FLT3, MFGE8, IFNGR1, IL1RAP, CXCR4, TXNIP, TFRC, CD1D, CCND3, MAP2K1, HMGB1; downregulated genes: CLEC4C, TLR7, LTB, IL21R, IFIH1, PIK3CD |
[76] |
| 40 BC (23 TNBC and 17 luminal (ER+/PR+/HER2-)) |
Human Inflammatory Cytokines and Receptors PCR Array, 84 genes | Downregulated in TNBC: interleukins (IL13, IL16, IL17C (IL21), IL17F, IL1A, IL3), interleukin receptor (IL5RA), cytokines (CSF2, OSM, TNSF13), chemokine (CCL26); upregulated in TNBC: interleukin receptor (IL10RB), chemokine (CXCL13), cytokine (IFNA2) |
[12] |
| 30 BC (18 TNBC and 12 ER-/PR-/HER2+) |
Human Breast Cancer PCR Array, 84 genes | Downregulated in TNBC: ERBB2, RASSF1, CDH1, MKI67, GATA3, GLI1, SFN, PTGS2, JUN, NOTCH1, CTNNB1, KRT8, SRC, and HIC1; upregulated in TNBC: CCNA1 |
[11] |
| 70 BC (8 TNBC, 5 luminal A, 20 luminal B, 19 luminal B-like, 13 HER2, 5 unknown) and 50 healthy controls |
qRT-PCR | Upregulated in TNBC compared to non-TNBC subtypes: VEGFR3 and PLXNA1 (co-receptors of NRP-1) | [102] |
2. PBMC Transcriptome of TNBC (ER-/PR-/HER2-) versus Hormone-Dependent BC (ER+/PR+/HER2-)
Foulds et al. profiled 770 immune-related genes in 14 TNBC and 9 luminal-A breast cancer patients [76]. Reminiscent of other studies, the immune transcriptome revealed a subgroup of three TNBC patients that had a unique immune expression profile, showing differential expression of genes functionally related to inflammation, including CD163 (a scavenger receptor involved in the resolution of inflammation), cytokine receptors such as IFNGR1 (CD119), IL21R, IL1R2, FLT3, IL1RAP, TXNIP (which promotes anti-inflammatory macrophages), and HMGB1 (which is upregulated under inflammatory conditions). Moreover, the combined low tissue expression of CD163 and CXCR4 and high tissue expression of THBS1 significantly correlated with a high risk of recurrence and poor survival rate in TNBC [76]. The authors suggested that the emergence of a distinct PBMC immune transcriptome subgroup may have implications for clinical decisions.
Balacescu et al. studied the transcriptome of nucleated blood cells from 29 treatment-naïve BC patients with invasive ductal carcinoma and seven healthy controls. The microarray analysis delineated two distinct clusters corresponding to BC and controls [5]. However, within the BC cluster, which included 14 TNBC (ER-/PR-/HER2-) and 15 hormone-dependent (ER+/PR+/HER2-) patients, the expression patterns were mixed and did not cluster according to the ER/PR status, in line with reports by other researchers. Even so, the ER-/PR-/HER2- and ER+/PR+/HER2- groups did show some individual differences in molecular profiles, and a specific 34-gene signature for TNBC, which was able to distinguish TNBC from both hormone-dependent patients and healthy controls, was identified [5]. The nucleated blood cell transcriptome indicated a strong involvement of tumor-related inflammation and immune response in TNBC. Pathway analysis revealed that TNBC was associated with altered systemic immune-related pathways, including chemokine signaling, IL-8 signaling, and communication between innate and adaptive immune cells. Importantly, this dysregulation correlated with increased inflammation and necrosis in the primary tumors. IPA upstream regulator analysis predicted AREG as the upstream regulator in TNBC (with downstream targets AREG/AREGB, CXCR4, EGR1, FOS, PLAU, and PTGS2), while two upstream regulators (AREG and F7) were predicted for hormone-dependent BC. Of note, since the modulation of systemic immune-related pathways was detected in TNBC, the authors concluded that immunotherapy may be a synergistic approach to chemotherapy for this cancer type, which is in line with the findings from other researchers. For example, Suzuki et al. assessed that gene expression profiling in PBMCs could help to obtain an immunological insight with clinical utility for BC—it could be useful for observing the immune reaction related to cancer progression and for monitoring the efficacy of immune-related cancer therapy [89].
Immunological profiling of 84 inflammatory molecules and their receptors in PBMCs of treatment-naïve ductal breast carcinoma patients, including 23 TNBC (ER-/PR-/HER2-) and 17 Her2-luminal (ER+/PR+/HER2-), revealed that TNBC patients had altered expression of PBMC genes associated with immunological status and presented with lower counts of lymphocytes and eosinophils than the ER+/PR+/HER2- patients [12]. Downregulated PBMC genes in TNBC included interleukins (IL13, IL16, IL17C, IL17F, IL1A, IL3), the interleukin receptor IL5RA, cytokines (CSF2, OSM, TNSF13) and the chemokine CCL26. On the other hand, interleukin receptor IL10RB, the chemokine CXCL13 and the cytokine IFNA2 were upregulated in TNBC. IFNA2 and IFN-alpha upregulation suggested the activation of downstream signaling cascade through IL10RB receptors. Overall, the hormone-receptor (ER/PR) expression on HER2-negative breast tumors was associated with distinct systemic PBMC-associated cytokine profiles.
In addition to the link between tumor hormone-receptor expression and the systemic cytokine response, the immune activity at the tumor site is also associated with the systemic response (SR). While a high inflammatory SR is observed in both ER-/HER2- and ER+/luminal B patients, systemic inflammation is associated with immune activity at the tumor site depending on subtype. Specifically, ER-/HER2- patients with low immune activity at the tumor site show a high inflammatory SR, while ER+/luminal B patients with high immune activity at the tumor site have a high inflammatory SR [85].
3. PBMC Transcriptome of TNBC (ER-/PR-/HER2-) versus Her2-Overexpressing BC (ER-/PR-/HER2+)
Tudoran et al. studied whether expression of HER2 on hormone-receptor-negative tumor cells influences the transcriptome of nucleated PBCs. An investigation of 84 breast-cancer associated genes in 18 TNBC (ER-/PR-/HER2-) and 12 Her2-overexpressing (ER-/PR-/HER2+) BC patients revealed 15 genes that were differentially expressed, even though HER2- and HER2+ groups had comparable counts of nucleated cells in blood [11]. Fourteen genes were downregulated in TNBC, including cell cycle regulators (CCNA1, JUN, MKI67, RASSF1, SFN), cell adhesion molecules (CDH1, CTNNB1, HER2), transcription factors (CTNNB1, GATA3, HIC1, JUN, NOTCH1) and signal transducer GLI1. On the other hand, the cell cycle regulator cyclin A1 was upregulated in TNBC [11]. Network analysis indicated that these genes are interconnected, regulate each other, and participate in cancer progression and the modulation of immune signaling. Since fine-tuned engagement of immune responses is needed in favorable treatment response, the altered immune signaling in peripheral blood cells of TNBC patients may contribute to their low treatment response rates, and the authors suggested that baseline monitoring of the immune status may help in treatment response prediction. Interestingly, although HER2-negative expression is characteristic of TNBC tumor cells, decreased HER2 expression was also observed on white blood cells from TNBC patients, in line with previous reports that suggested a correlation between tumor and blood HER2 expression levels [11,100,101].
4. Peripheral Blood Cell Transcriptome and Therapy Response and Prognosis in TNBC Patients
The identification of patient immune pathways that are specifically altered in individual BC subtypes can potentially lead to facilitating the selection of the optimal treatment. This holds implications for TNBC, known for treatment nonresponse and poor prognosis.
Recent work has identified NRP-1 (a non-tyrosine kinase receptor) and its interacting molecules as possible drug targets and as biomarkers for predicting poor prognosis in BC [102,103,104]. NRP-1 is involved in primary immune response initiation by mediating the interaction between dendritic cells and resting T cells [105]. Moreover, accumulating evidence indicates that NRP-1 has a role in promoting cancer due to its involvement in the evasion of immune surveillance [106]. Tumor tissue NRP-1 and its soluble isoforms in plasma are upregulated in advanced nodal and metastatic BC, and tumor tissue NRP-1 expression is increased in TNBC compared to other subtypes. In contrast, in PBMCs of BC patients, a downregulation of NRP-1 and its interacting molecules SEMA4A and SNAI1 is seen [102]. This decrease in BC compared to healthy subjects suggests that the interaction of these molecules on PBMCs may not participate in tumor immune evasion but rather may play a protective role against BC. This is supported by evidence that SEMA4A and SNAI1 expression on PBMCs declines with increased tumor size and overall disease stage. Importantly, PBMC upregulation of VEGFR3 and PLXNA (i.e., co-receptors of NRP-1) can distinguish TNBC from other BC subtypes, suggesting their expression on PBMCs may have potential to determine BC cases susceptible to immunotherapy [102]. Altogether, the differences in plasma, tissue and PBMC profiles suggest that NRP-1 has multiple cell type-specific roles in BC, being a risk factor in plasma and tumor tissue and a protective factor in PBMCs [102]. However, Suzuki et al. reported that NRP-1 was elevated in PBMCs of ER-HER2- patients in the “monocyte-activating” immunological subgroup, and therefore further elucidation of NRP-1 in BC is warranted [89].
Neoadjuvant chemotherapy (NAC) is the standard care for a subset of BC patients. Recent advances in immunotherapy in combination with chemotherapy for TNBC have brought forward the need to understand how chemotherapy influences local and systemic immune responses. Axelrod et al. reported that during chemotherapy, increased expression of several immune-related genes and groups of genes (particularly related to T-cells) in the tumor tissue is associated with improved prognosis in TNBC, but not other BC subtypes [107]. Moreover, in peripheral blood of TNBC patients after NAC therapy, high expression of cytolytic markers was detected by single-cell RNA sequencing of the CD8+ PD-1HI population (enriched in tumor-reactive T-cells), and a substantial increase in cytokines expressed by this population was measured [107]. The gene expression signature of immune activation and cytotoxicity (FGFBP2 + GNLY + GZMB + GZMH + NKG7 + LAG3 + PDCD1 − HLA-G) in CD8+ PD-1HI T-cells of TNBC patients and in post-NAC whole blood of BC patients was associated with persistent disease following chemotherapy and disease recurrence after surgery [107,108]. These findings highlight that local and systemic immune-related signatures may help to identify patients with good prognosis following NAC and those likely to benefit from additional immunotherapeutic approaches.
Gene expression is strongly influenced by DNA methylation, an epigenetic modification associated with transcriptional silencing. Tumor suppressor genes (e.g., BRCA1, ATM) are often hypermethylated in patient tumors and in peripheral blood [109,110]. Targeted and genome-wide DNA methylation studies have identified specific methylation changes associated with increased breast cancer risk and BC prognosis [111]. For instance, BRCA1 promoter methylation in peripheral blood was found to correlate with an increased risk of developing TNBC (but not ER+ BC) and was also associated with high-grade tumors [110]. Interestingly, BRCA1 promoter methylation in PBCs correlated with an increased risk of developing BC even in non-carriers of BRCA1 mutation [112,113]. Methylation is also important during cancer treatment, where it is linked with drug resistance. Treatment with chemotherapeutics or immunotherapy was shown to cause acquired methylation-associated resistance in cell models, and therapy resistance was also observed in BC patients [114,115]. The use of epigenetic drugs (e.g., DNMT inhibitors) is being tested in combination with chemotherapy, immunotherapy, and other treatments to overcome epigenetic alterations and treatment resistance (Figure 2) [109,116].
This entry is adapted from the peer-reviewed paper 10.3390/cancers14030591
This entry is offline, you can click here to edit this entry!
