Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Engineering, Biomedical
Due to superior biocompatibility, thermal stability, and alternative functionalization, deoxyribonucleic acid (DNA) is becoming a fascinating biological material used for biosensing. It is widely acknowledged that DNA and its assembly structure can be applied for detecting specific targets, including nucleic acids, proteins, metal ions, and small biological molecules. With the development of DNA nanotechnology, dynamic networks based on DNA hybridization can be used to amplify the signals of biosensors.
- DNA-based biosensors
- DNA aptamer
- DNAzyme
- DNA origami
- DNA nanotechnology
1. Introduction
Due to superior biocompatibility [1], thermal stability [2,3,4,5], and alternative functionalization [6,7,8], deoxyribonucleic acid (DNA) is becoming a fascinating biological material used for biosensing. It is widely acknowledged that DNA and its assembly structure can be applied for detecting specific targets, including nucleic acids, proteins, metal ions, and small biological molecules [9,10,11,12,13,14,15]. With the development of DNA nanotechnology, dynamic networks based on DNA hybridization can be used to amplify the signals of biosensors. In addition, DNA is also a powerful material to assemble complex 3D nanostructures and organize the other functional units.
Compared to commonly used bioprobes, more durable biological activity, remarkable addressability, and adjustable rigidity make DNA a promising candidate for intelligent biosensing. It has been reported that through manual screening and modification, DNA probes, like aptamer, have better thermal stability [16,17,18], adjustable biological affinity [19,20,21], and higher resistance to nucleases enzyme attack [22,23]. DNA can also be used to build programmable supermolecule structures as the template to realize the precise controlling of the spatial position of the modifications, which could significantly improve the performance of the biosensor and even inspire researchers to propose novel biosensors [24,25,26].
Based on the great potential of DNA biosensors. A large number of reports have reviewed the basic principles and the recent advances of the biosensors based on DNA aptamers [27,28,29,30,31], DNAzyme [32,33,34,35], DNA hairpins [36,37,38,39], DNA tiles [40,41,42], and DNA origami [43,44,45,46,47]. Although these reports have introduced DNA biosensors from different aspects in detail, a comprehensive overview of DNA-based biosensors is still needed.
2. Functional DNA Strands-Based Biosensors
Biosensors rely on the interaction of a molecular probe with a specific affinity to the target analyte [54]. In the past few decades, enzymes [55], antibodies [56,57], and oligonucleotides [58,59] have been widely used as biosensor probes with specific recognition functions. Compared with the biosensors based on an enzyme or antibody, biosensors based on DNA probes have advantages of high thermal tolerance, easy modification, and efficient surface regeneration because of the stable chemistry [60]. More significantly, DNA probes with different affinity for target analytes can be obtained through directed screening of DNA libraries [61,62]. In this section, biosensors based on two kinds of functional DNA strands, DNA aptamer, and DNAzyme are introduced.
2.1. DNA Aptamer Biosensors
Aptamer refers to a series of synthetic nucleic acids capable of binding to a specific target. The first aptamer was obtained in 1990 by Ellington et al. [63]. Compared with the traditional bioprobes like the antibody, DNA aptamers could better adapt to extremely high temperatures, pH values, and high ionic concentrations. Furthermore, a DNA aptamer allows much simpler modification of functional groups without losing biological activity. Additionally, the manufacturing cost is also dramatically reduced with the rapid development of DNA synthesis technology. All these advantages facilitate the wide application of DNA aptamers in various biosensors.
The most used strategy to detect a biotarget with a DNA aptamer is to functionalize the DNA aptamer with a report molecule (ferrocene, methylene blue) and an immobilization molecule (alkane thiol, alkane amino, streptavidin, and hydrazoate) at the 5′ end and 3′ end of the DNA strand, respectively. The change of DNA aptamer construction could be read out by detecting the electrochemical change on the electrode surface. Liu et al. developed an electrochemical sensor based on a 34-mer IFN-γ-binding aptamer for interferon-gamma (IFN-γ) detection [64] (Figure 2A). In their study, the proposed DNA aptamer was fixed on the surface of the electrode through a covalent reaction of gold and alkyl mercaptan. When there was no specific target, the DNA aptamer self-folded to form a secondary spatial loop structure, which caused the reporting molecules to contact the sensor electrode surface. The combination of the target and the DNA aptamer changed the aptamer’s conformation, enlarging the distance between the reporting molecule and the electrode surface. This led to the change of electronic transfer efficiency between the reporting molecules and the electrode surfaces. The limit of detection (LOD) reached 0.06 nM, and the linear detection range was extended to 10 nM. In a similar study (Figure 2B), Chen et al. developed an electrochemical sensor array based on the 34-mer IFN-γ binding aptamer modified with MB and disulfide, using standard semiconductor processes [65]. This sensor could detect IFN-γ ranging from 1 to 500 ng/mL with a LOD of 1.3 ng/mL.
A DNA aptamer can also be decorated with conjugated polymers, which has been widely applied as the report tags in fluorescent and colorimetric biosensors because of its excellent optoelectrical properties [66]. The DNA aptamer can absorb onto the conjugated polymers through the electrostatic force [67]. When binding with the biotarget, the conformation change of the DNA aptamer will lead to the adjustment of the conjugated polymer’s constructure, which will influence the absorption and emission wavelength of the conjugated polymers [68]. This method has been successfully used to detect human α-thrombin, with a LOD of 2 × 10−15 M [69].
Although functional DNA aptamer biosensors exhibit many advantages, a complex modification process is needed to immobilize the aptamers onto the sensor surface firmly. Graphene oxide (GO), which shows excellent photoelectric properties and carrier transport capability, is a widely used 2D material to simplify the process [70,71,72,73,74,75]. It has been reported that single DNA strands could be tightly adsorbed on the GO surface through base-stacking and hydrogen bonding without chemical modification [76]. Yu et al. proposed a simple method to detect microscale Pb2+ using electrochemical aptamer sensors modified with electrochemically reduced graphene oxide (ERGO) [77]. In their work (shown in Figure 2C), the ERGO was deposited on the glassy carbon electrode, and the guanine-rich DNA aptamer modified with methylene blue (MB-aptamer) was physically adsorbed onto the ERGO through the π-π interaction. The Pb2+ led the MB-aptamer to fold to G-quadruplex and separate from the electrode surface, which could weaken the electrochemical signal of the sensor surface. This strategy had a linear range from 10−5 to 10−9 M and a LOD of 0.51 fM.
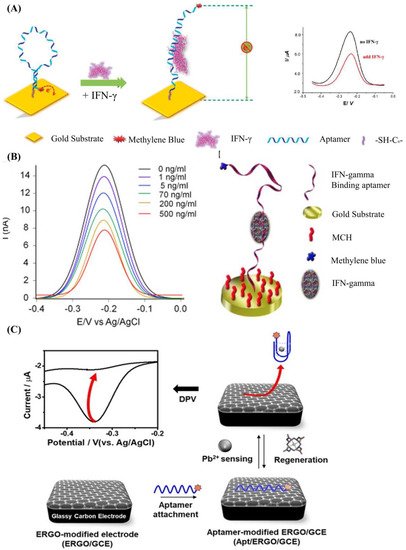
Figure 2. (A) Schematic of aptamer-based electrochemical sensor for IFN-γ. Reprinted (adapted) with permission from [64]. Copyright (2010) American Chemical Society. (B) Demonstration of the MB-tagged aptamer modified electrode. Reprinted (adapted) with permission from [65]. Copyright (2014) Elsevier. (C) Schematic diagram depicting the fabrication of Ap/ERGO/GCE-based electrochemical aptasensor for the detection of Pb2+. Reprinted (adapted) from [77]. Copyright (2019) MDPI, Basel, Switzerland, under the Creative Commons Attribution License.
For DNA aptamer biosensors, the dynamic range and sensitivity, which are limited by the Langmuir isothermal adsorption model, are also not flexible enough to suit the different ranges of detection concentration required [78,79]. As shown in Figure 3A, aptamers with different detection ranges can be mixed to achieve a wider detection range. This method is quite suitable for the detection of several viruses [80] and inflammatory biomarkers, the detection ranges of which may span several orders of magnitude [81]. However, due to the unchanged maximum response of the sensor, the sensitivity of this kind of biosensor will be decreased. Figure 3B shows that the detection range can also be changed by adjusting the aptamers’ conformation, which shifts the detection range of the aptamer rather than expands it. Therefore, its biosensing sensitivity will not be decreased, which is suitable for applications like the detection of cancer biomarkers [82] (high sensitivity instead of a wide detection range is preferred).
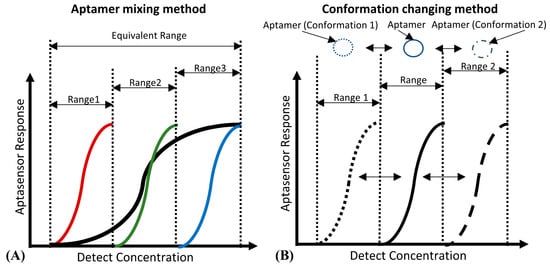
Figure 3. (A) The principle of the “aptamer mixing method”. It can be seen that the detection ranges of aptamers, which have a high affinity (marked in red), medium affinity (marked in green) and low affinity (marked in blue) to the same biotarget, are narrow. However, the equivalent detection range of their mixture is wide. (B) The principle of the “conformation changing method”. By inducing the aptamer to change to the conformation with high affinity (Conformation 1), and the conformation with low affinity (Conformation 2), the detection range of the aptamer can shift to the low detection concentration area (Range 1) and the high detection concentration area (Range 2) respectively.
In addition to improving the biosensing performance, the cost of these biosensors should be taken into consideration, which indicates that it is meaningful to develop reusable biosensors. One commonly used way is to flush the sensor surface with running buffer rapidly for a long time to recover the sensor surface. A prominent drawback is the baseline of the sensor may drift after flushing the sensor surface, which can influence the detection range of the sensor. Moreover, the overly fast flow will physically damage the modification of the sensor surface, especially when the aptamers are immobilized onto the sensor through electrostatic force. Another way to recover the sensor surface is to use a denaturant, a chaotropic, or surfactants. However, these reagents are harmful to the bioactivity of the sensor. To overcome the above-mentioned disadvantages, a method called DNA aptamer substitution was proposed to realize the recovery of the sensor surface more effectively and more simply [83]. Compared to the two methods above, there was no residual biotarget on the sensor surface, and this recovery process did not change the surface distribution of the DNA aptamer.
2.2. DNAzyme Biosensors
Enzymes, which have high catalytic efficiency, are widely used in biosensing [84,85,86,87]. However, the enzymatic activity can be affected by many environmental factors, which limit the application in biosensors [88,89]. It is found that some manually screened DNA strands which are more adaptive to the environment and resistive to the nuclease degradation also have enzymatic activity [32,90]. These DNA strands, named DNAzyme, have excellent potential to be efficient biometric probes. Among the DNAzymes with different functions, the nucleic acid cleavage function and the catalytic function of peroxides are widely used in biosensing.
The DNAzyme with nucleic acid cleavage function consists of a loop-shaped catalytic domain flanked by two substrate-recognition domains [91]. Figure 4A shows the basic principle of DNAzyme with nucleic acid cleavage function. The substrate-recognition domains can capture the substrate strand through the Watson–Crick model. The cleavage will not be activated without the existence of the catalytic core. When the external catalytic core embeds into the catalytic domain of the DNAzyme, the cleavage process is stimulated. The substrate strand is then cut off and separated from the substrate-recognition domains because the melting temperature decreases [92,93,94,95,96]. During this process, only certain small molecules (like metal ions [97,98,99] and amino acids [100]) can serve as the catalytic core of the corresponding DNAzyme. Therefore, the stimulation of the cleavage process is highly specific, which can be utilized for small-molecule detection.
The most commonly used method based on this principle is called the molecular beacon [101]. Generally, there are three types of molecule beacons: the beacon with a single quencher, the beacon with double quenchers, and the beacon with the hairpin-shaped substrate (shown in Figure 4B–D, respectively). The beacon with a single quencher refers to the molecule beacon modified with one fluorescence molecule and one quencher. Li et al. developed a highly sensitive DNAzyme biosensor based on the beacon with a single quencher to detect lead ions [102]. In this study, the fluorescence sensor was constructed by modifying the 5′ end of the substrate DNA (Rh-17DS) with fluorophore 6-carboxylic tetramethylrhodamine (TMR) and modifying the 3′ end of the DNAzyme chain (17E-Dy) with fluorescence quencher 4-(4′-dimethylaminophenylazo) benzoic acid (Dabcyl). The Pb2+ activated the DNAzyme’s cleavage process, leading to the separation of the fluorescence molecule (TMR) and the quencher (Dabcyl). Therefore, the FRET between the TMR and the Dabcyl was interrupted, and the intensity of the fluorescence was enhanced. Their proposed sensor had an 80-times-higher response for Pb2+ than for the other divalent metal ions. However, this biosensor only worked well at low temperatures. When the ambient temperature exceeded the melting temperature, some of the DNA enzymes would release the substrate strand without cleaving, which would cause high background noise and weaken the selectivity of the method. To solve this issue, the beacon with double quenchers, which refers to the beacon modified with one fluorescence molecule and double quenchers, was proposed. Liu et al. proposed an improved fluorescent DNAzyme sensor based on the beacon with double quenchers [103]. As shown in Figure 4C, the fluorescent molecule (FAM) was quenched not only by the quencher (Dabcyl) on the DNAzyme (17E-Dy) but also by the quencher (Dabcyl) on the substrate strand (Rh-17DS). Even when the substrate strand was separated from the DNAzyme abnormally, this clever design could still keep the fluorescent molecules in a quenching state, because the Dabcyl on the substrate strand could still quench the FAM. Their results showed that the proposed sensor’s relative fluorescence intensity was increased by 60%. The beacon with hairpin-shaped substrate refers to the stem-loop of the DNA hairpin that serves as the substrate of the DNAzyme. Zhao et al. developed a DNAzyme-based amplified biosensor [104]. As shown in Figure 4D, the fluorescent-quench pair was modified at the DNA hairpin’s end. The cleavage site was set on the loop of the hairpin. Induced by zinc ions, the DNAzyme cut the loop and released fragments of nucleic acid with fluorescent-quenching molecules to increase the intensity of the fluorescence. This strategy did not need to modify DNAzyme, avoiding the decrease of the DNAzyme’s catalytic activity.
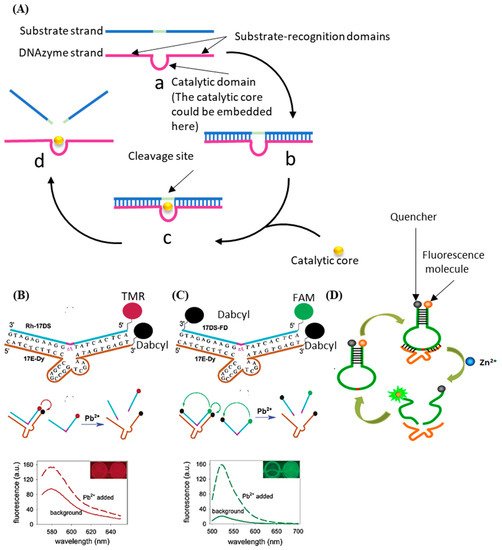
Figure 4. (A) The basic principle of the DNAzyme with nucleic acid cleavage function. (B) Schematic diagram of previous DNAzyme-based Pb2+ sensor. (C) Schematic diagram of improved DNAzyme-based Pb2+ sensor. Reprinted (adapted) with permission from [103]. Copyright (2003) American Chemical Society. (D) Schematic diagram of DNAzyme-based amplified biosensor. Reprinted (adapted) with permission from [104]. Copyright (2013) American Chemical Society.
DNAzyme with the catalytic activity of peroxides was firstly discovered by Sen et al. in 1998 [105]. As shown in Figure 5A, in the presence of potassium ions, DNA sequences rich in guanine can combine with hemin to form HRP-DNAzyme, which could catalyze 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS2−) or tetramethylbenzidine (TMB) to the colored products. HRP-DNAzymes are commonly applied as report tags of DNAzyme and DNA aptamer biosensors because HRP-DNAzyme can produce intense fluorescent or colorimetric signal and will not largely affect the bioactivity of these DNA biosensors. Willner et al. designed an aptamer-HRP-DNAzyme hairpin biosensing structure for detecting AMP and lysozyme [106]. In their research (shown in Figure 5B), the hybridization of the analyte with the DNA aptamer sequence causes the DNAzyme to untie from the stem and form a G-quadruplex structure. Catalyzing the ABTS2− to the colored ABTS●− by the HRP-DNAzyme, an amplified optical signal could be obtained at the same time for detecting respective analytes. HRP-DNAzyme can also catalyze the polymerization of aniline to form polyaniline, which shows superior SPR signal-enhancing ability. Based on this, Li et al. proposed an SPR biosensor based on HRP-DNAzyme for signal amplification [107]. In the study, bleomycin was used as the target, and a much lower LOD down to 0.35 pM was realized.
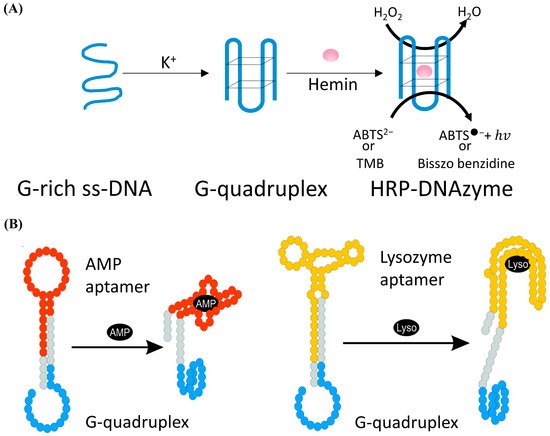
Figure 5. (A) Schematic of the form and the function of the HRP-DNAzyme. (B) Schematic analysis of adenosine monophosphate (AMP) or lysozyme (Lyso) by the aptamer—DNAzyme hairpin structure; reprinted (adapted) with permission from [106]. Copyright (2009) American Chemical Society.
3. DNA Hybridization-Based Biosensors
Rapid and sensitive detection of specific biomarkers is of great importance in biochemical analysis, especially in the global pandemic of COVID-19 nowadays [108,109,110]. At present, the widely used nucleic acid detection methods include polymerase chain reaction (PCR) [111,112], loop-mediated thermal amplification (LAMP) [113], DNA chip-based microarray [114], and enzyme-linked immunosorbent assay (ELISA) [115]. However, these methods require complex testing equipment, professionally trained inspectors, long incubating time, and complex manufacturing processes. These shortcomings limit the application of these methods in airports, train stations, communities, etc. The detection technique based on DNA hybridization amplification, which is of high detection speed, sensitivity, and stability, shows great potential to overcome the above-mentioned drawbacks. In this session, nucleic acid hybridization-based biosensors, including the DNA hairpin biosensors, the HCR biosensors, and the CHA biosensors used to detect specific biology targets, were introduced.
3.1. Biosensor Probes Based on DNA Hairpin
DNA hairpin refers to the hairpin structure formed by ssDNA with a self-complementary sequence. Based on Watson-Crick’s pairing principle, this structure shows high specificity and can convert the hybridization to the physical signal easily. Thus, the DNA hairpin-based probe can be a powerful tool for detecting target nucleic acid fragments. Fang et al. proposed a kind of molecular beacon for surface-immobilized DNA hybridization studies [116]. In their research, the hairpin was modified with tetramethylrhodamine (TMR) and dimethylaminoazobenzen aminoexal-3-acryinido (DBCAL) as fluorescence–quenching pairs. TMR and DBCAL were detached with the opening of the hairpin by the target, which led to the increase of the fluorescence signal. This sensitivity could reach as low as the nanomolar scale. Fan et al. developed an electrochemical DNA (E-DNA) biosensor based on the DNA hairpin to detect the sequence-specific DNA [117]. In the study, a DNA hairpin, which was modified with methylene blue (MB) and a ferrocene molecule, was immobilized onto the working electrode through the covalent reaction between gold and hydro sulphonyl. As the sequence-specific DNA opened the hairpin and caused the detachment of MB from the sensor surface, the electrochemical signal of the sensor decreased. The LOD of this sensor could reach as low as 10 pM. Compared with the fluorescence method, this biosensor did not need bulky equipment and was not affected by the sample’s light transmittance. However, the response current was inversely proportional to the target molecule’s concentration, which indicated that the sensor response under high concentration might be drowned in background noise. One method to overcome the drawback is designing a hairpin structure based on “signal gain mode” [118]. Another method is introducing a reference probe to reduce the background noise [119].
3.2. Signal-Enhanced Biosensors Based on DNA Hybridization
Although the biosensors based on DNA probes like aptamers, DNAzymes, or DNA hairpins have achieved high sensitivity, chemical stability, and low manufacturing costs, the LOD is still limited by the surface density of DNA probes. Denser surface distribution of DNA biometric elements could theoretically increase the probability of combining the target and sensor surface probe [120]. However, the steric hindrance effect caused by the over-density of DNA probes on the sensor surface prevents the target from binding to the immobilized probes.
The cyclic amplification techniques based on hairpins, which mainly include HCR and CHA, are developed to enhance the biosensing signal largely. HCR was triggered by an initial single strand of nucleic acid, and the DNA hairpins with sticky ends alternately hybridized to form a long double helix. Hou et al. developed an HCR electrochemical sensor based on the signal attenuated mode for detecting micro-RNA with high selectivity [121]. The sensor showed good sensitivity with a LOD of 1 pM. HCR may be triggered without the existence of the target and cannot be stopped automatically, while CHA needs the participation of original ss-DNA as the trigger in every reaction round, which makes the biosensors based on CHA more robust. Duan et al. proposed a fiber optic biosensor which was based on CHA and nanocomposites-assisted signal amplification to detect 18 samples of food [122]. The LOD of the sensor could reach 12 pM. In addition, researchers are trying to combine CHA with other advanced biomedical analysis methods. Wu et al. combined CHA with rolling circle amplification (RCA) for the telomerase activity detection with high sensitivity both in vitro and in situ [123]. As shown in Figure 6, the RCA step was used to monitor the activity of telomerase and amplify the target, and the CHA step was used to convert the increase of the RCA product into fluorescence enhancement to readout.
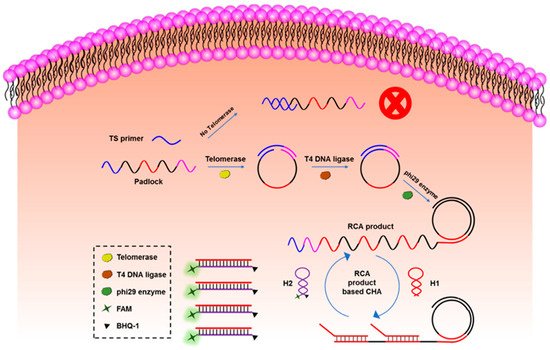
Figure 6. The principle of CHA-assisted RCA for telomerase activity detection. Reprinted (adapted) with permission from [123]. Copyright (2020) American Chemical Society.
This entry is adapted from the peer-reviewed paper 10.3390/bios12030183
This entry is offline, you can click here to edit this entry!
