This is a summary of the publication (https://doi.org/10.3390/plants9101272) where the complete coding sequence of olive leaf yellowing-associated virus (OLYaV) has been determined and the phylogenetic relationships with other members of the Closteroviridae family were analyzed. New insights into the taxonomy of the family were achieved suggesting that OLYaV, persimmon virus B and actidinia virus 1, could represent a new genus in the family (Olivavirus).
- Closteroviridae
- Thaumatin-like protein
1. Introduction
Closteroviruses are composed by one, two or three positive-sense, single-stranded RNA segments with lengths between 13,000 to nearly 19,000 nt[1], and the family Closteroviridae is currently divided in four genera, Closterovirus, Ampelovirus, Velarivirus and Crinivirus, which share common features in the genome organization: i) the presence of ORF1ab that includes one or two papain-like leader proteases, conserved domains for methyltransferase and helicase and an RNA-dependent RNA polymerase expressed through +1 ribosomal frameshift; ii) the existence of an HSP70 homologue gene; iii) an HSP90 homologue gene; iv) a capsid protein (CP) and v) in most members, the presence of a diverged, duplicate copy of the capsid protein, the minor capsid protein (CPm). The genus demarcation criteria in the family is based on the phylogenetic relationships in the RdRp, CP, HSP70h aa sequences, vectors, number of genomic RNAs, number and organization of ORFs and virion length[1]. Seven species in the family remain unassigned to a genus: Blueberry virus A (BVA), Actidinia virus 1 (AV1) and Persimmon virus B, for which complete genomes are available; Mint vein banding-associated virus (MVBaV) and OLYaV, for which only partial genomic sequences are available; and Alligatorweed stunting virus and Megakepasma mosaic virus without available nucleotide sequences[1]. Among these species OLYaV has a closest relationship with PVB [2] and AV1[3] based on the phylogenetic analysis of the partial OLYaV genomic sequences. PVB and AV1 lack the minor CP (CPm) and code for a thaumatin-like protein, unreported for other viruses of the family. In this study the full coding potential of OLYaV (isolate CS1) was obtained by high throughput sequencing (HTS) of total RNA from a 35 years-old olive tree (cv. Zarzaleña) from Brazil and the phylogenetic analysis of ORF1a, ORF1b (RdRp), HSP70, HSP90 and CP with their homologues in other representative species of the Closteroviridae family was performed opening new possibilities in the taxonomical classification of OLYaV.
2. Findings
2.1. Genomic organization of OLYaV
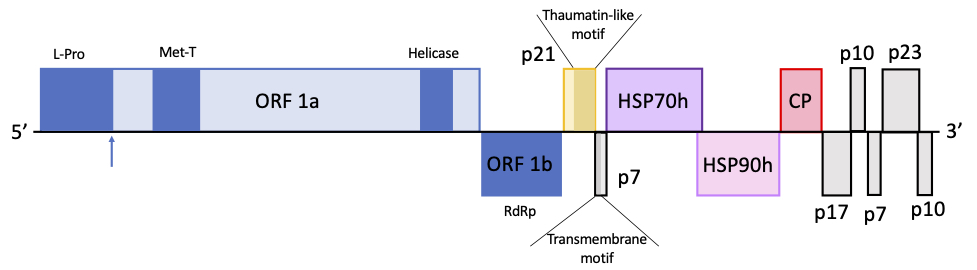
The genome structure of the isolate OLYaV-CS1 predicts 11 ORFs encoding proteins some of which have homologies with those of other Closteroviridae members (ORF 1a and b, ORF2-thaumatin-like protein, ORF4-HSP70h, ORF5-HSP90h and ORF6-CP) while others have no homologous counterpart in the GenBank database, ORF3-p7, ORF7-p17, ORF8-p10, ORF9-p7, ORF10-p23 and ORF11-p10 (Figure 1).
Figure 1. Genome organization of olive leaf yellowing-associated virus CS1 isolate. L-Pro: papain-like leader protease; Met-T viral methyltransferase domain; Helicase: viral helicase domain; RdRp: RNA dependent RNA polymerase. Blue arrow indicates tentative L-Pro cleavage site position.
2.2. Phylogenetic analysis of RdRp, HSP70 and CP
The phylogenetic analyses were performed using the best substitution model computed by MEGA X with the lowest BIC (Bayesian information criterion) and with 500 bootstraps (Figures 2-4). For all conserved proteins, the best substitution model was the Le and Gascuel one, employing the same equilibrium frequencies (+F) with rates among sites gamma distributed and with invariant sites (G+I). Representative isolates from each Closteroviridae species were included. Here we present the phylogenetic analysis of the three ORFs used for demarcation criteria. All common shared proteins among Closteroviridae members were also analyzed and can be visualize in the research paper from which is based this review (10.3390/plants9101272).
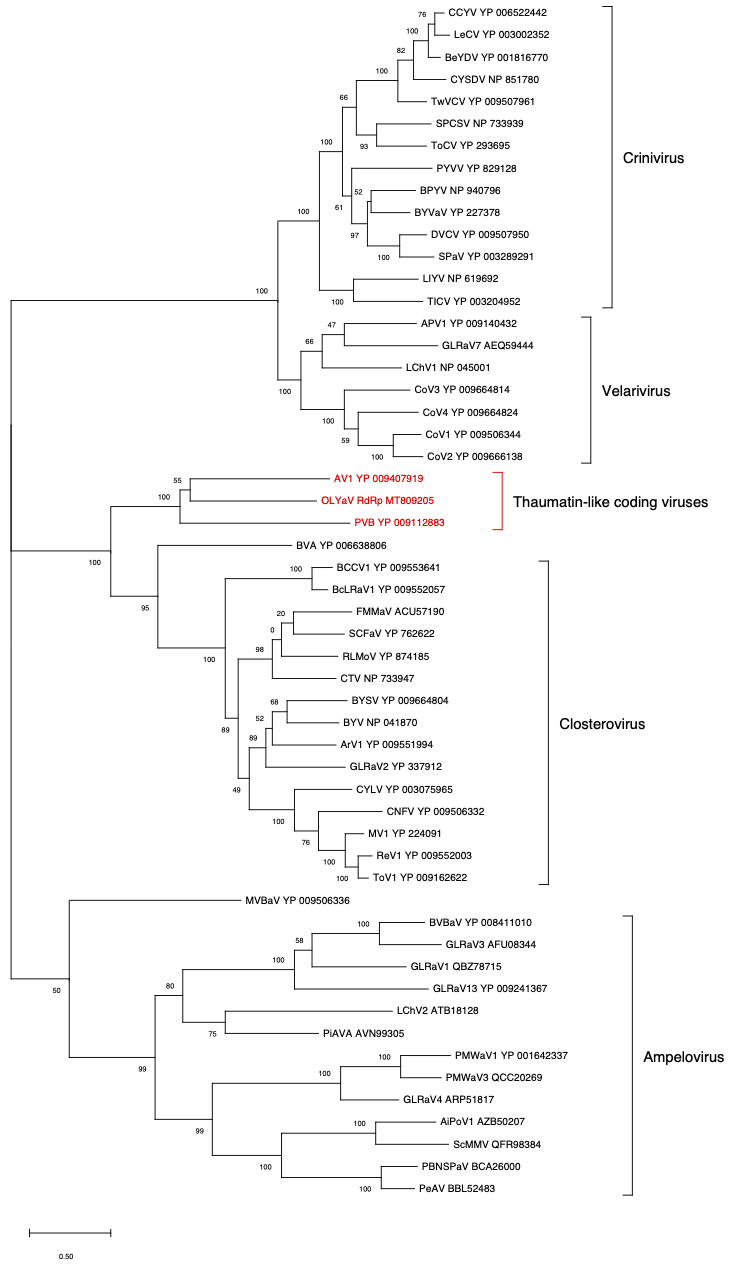
ORF1b (RdRp)
Figure 2. Maximum likelihood phylogenetic tree using the best substitution model by MEGA X of RdRp from representative members of the family Closteroviridae. Proteins of the isolates are identified by their protein_id number. The scale bar shows the genetic distance. Bootstrap percentages (500 re-samples) are indicated on the branches.
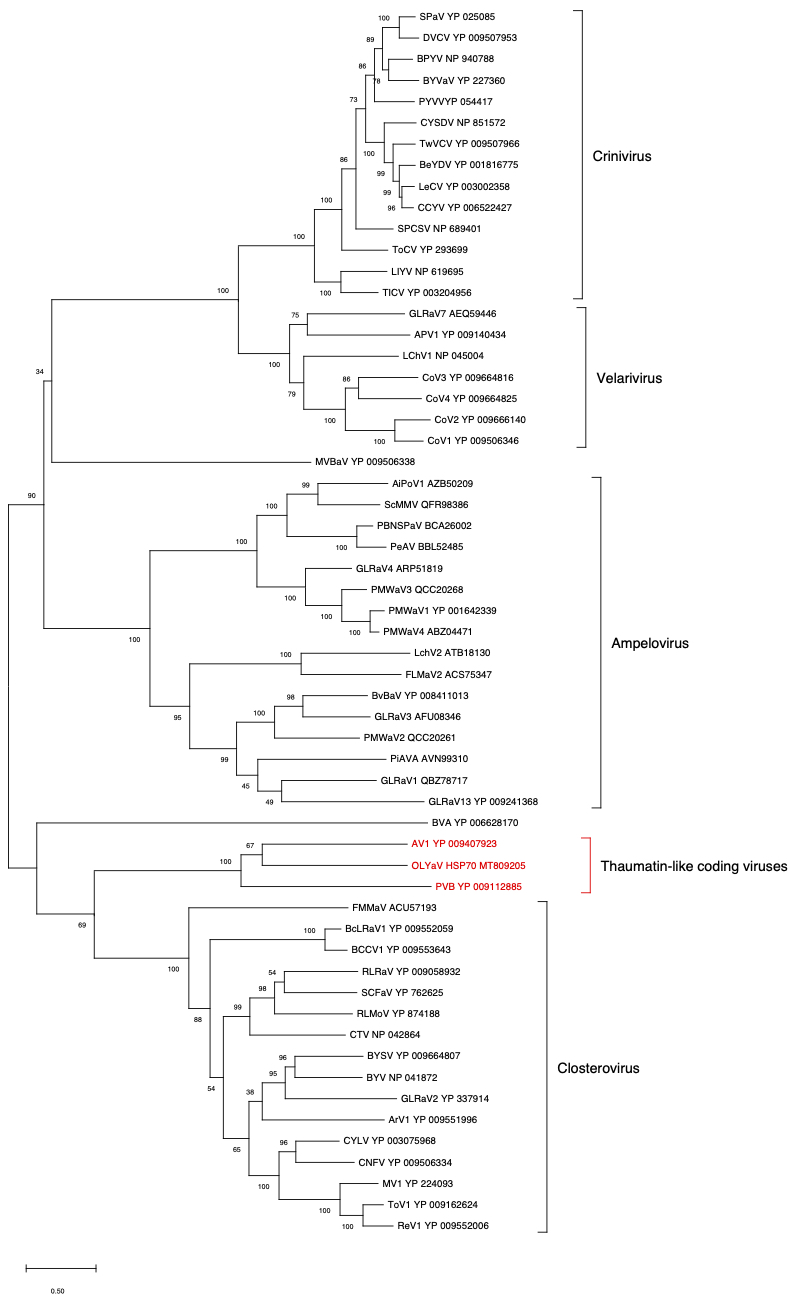
ORF4 (HSP70)
Figure 3. Maximum likelihood phylogenetic tree using the best substitution model by MEGA X of HSP70h from representative members of the family Closteroviridae. Proteins of the isolates are identified by their protein_id number. The scale bar shows the genetic distance. Bootstrap percentages (500 re-samples) are indicated on the branches.
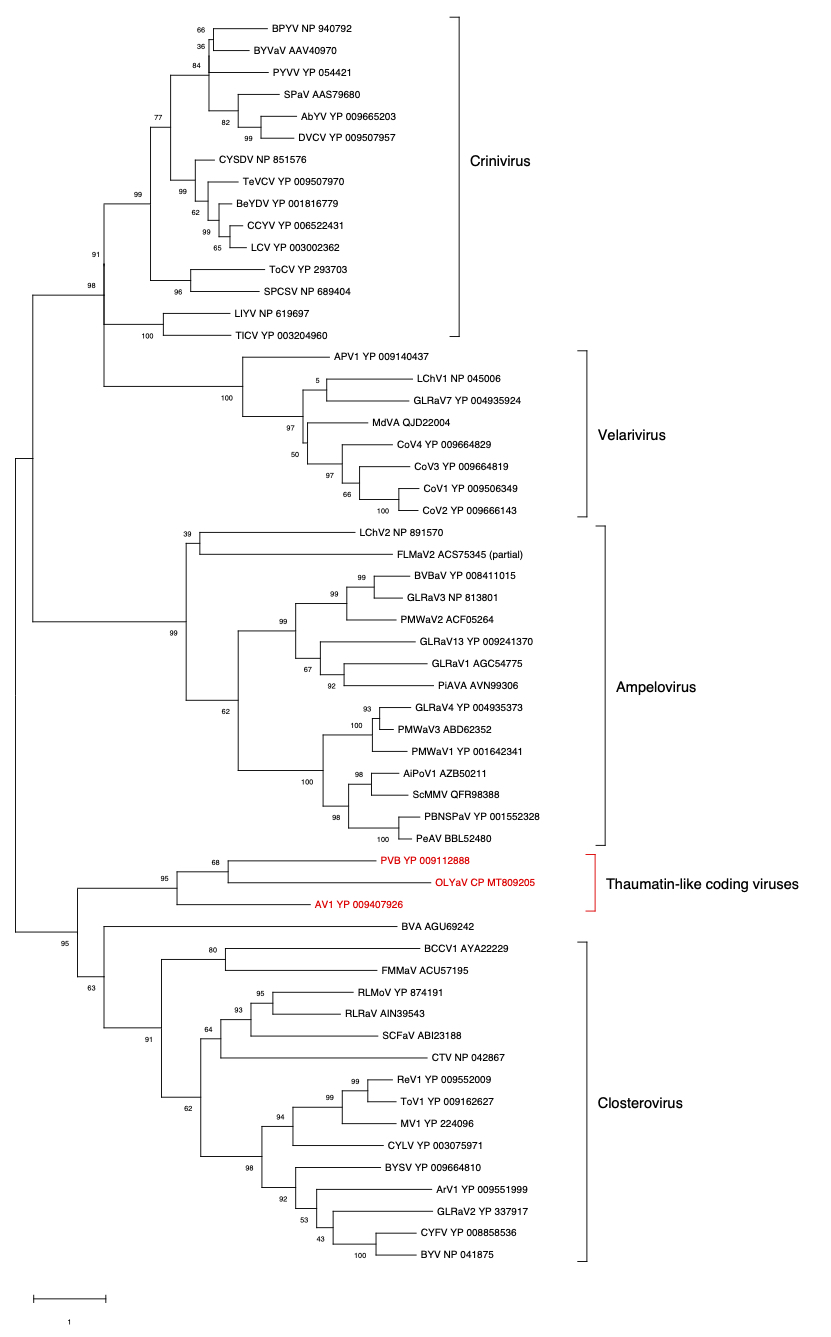
ORF6 (CP)
Figure 4. Maximum likelihood phylogenetic tree using the best substitution model by MEGA X of CP from representative members of the family Closteroviridae. Proteins of the isolates are identified by their protein_id number. The scale bar shows the genetic distance. Bootstrap percentages (500 re-samples) are indicated on the branches.
2.3. Other interesting findings
1) ORF1a codes for only one papain-like leader protease as other members of the family like BYV, MV1, LChV1 or LIYV[4].
2) There was a large deletion in the ORF1b of the Brazilian isolate as compared to the reference sequence, however it results in a larger RdRp for CS1 isolate (26 aa larger), because of the absence of the stop codon present in NC_043417.
3) ORF2 encodes for a thaumatin-like protein.
4) OLYaV does not code for a CPm protein.
Interestingly, 3) and 4) features are shared by two others unassigned Closteroviridae species, AV1 and PVB[2][3].
Conclusions
The genomic characerization of OLYaV, the discovering in the OLYaV genome of two genomic features shared by AV1 and PVB (the coding sequence for a thaumatin-like protein the lack of a CPm) and the confirmation of the close phylogenetic relationship between OLYaV, AV1 and PVB in all common shared Closteroviridae proteins, provide clear markers and strong support for the creation of a novel genus within the family to gather these three species.
This entry is adapted from the peer-reviewed paper 10.3390/plants9101272
References
- Fuchs, M., Bar-Joseph, M., Candresse, T., Maree, H.J., Martelli, G.P., Melzer, M.J., Menzel, W., Minafra, A., Sabanadzovic, S. and ICTV Report Consortium. ICTV Virus Taxonomy Profile: Closteroviridae, J Gen Virol 2020, 101, 364-365.
- Ito, T., Sato, A., Suzaki, K. An assemblage of divergent variants of a novel putative closterovirus from American persimmon. Virus Genes, 2015, 51, 105–111.
- Blouin, A.G., Biccheri, R., Khalifa, M.E., Pearson, M.N., Poggi Pollini, C., Hamiaux, C., Cohen, D., Ratti, C. Characterization of Actinidia virus 1, a new member of the family Closteroviridae encoding a thaumatin-like protein. Arch Virol, 2018, 163, 229–234.
- Peng, C.W., Peremyslov, V.V., Mushegian, A.R., Dawson, W.O., Dolja, V.V. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J Virol 2001, 75, 12153-12160.