Polycomb Repressive Complex 2 (PRC2) is arguably the best-known plant complex of the Polycomb Group (PcG) pathway, formed by a group of proteins that epigenetically represses gene expression. PRC2-mediated deposition of H3K27me3 has amply been studied in Arabidopsis and, more recently, data from other plant model species has also been published, allowing for an increasing knowledge of PRC2 activities and target genes. How PRC2 molecular functions are regulated and how PRC2 is recruited to discrete chromatin regions are questions that have brought more attention in recent years. A mechanism to modulate PRC2-mediated activity is through its interaction with other protein partners or accessory proteins. Current evidence for PRC2 interactors has demonstrated the complexity of its protein network and how far people are from fully understanding the impact of these interactions on the activities of PRC2 core subunits and on the formation of new PRC2 versions.
- PRC2
- chromatin
- protein interactors
- H3K27me3
- Arabidopsis
- transcription
- development
1. Background
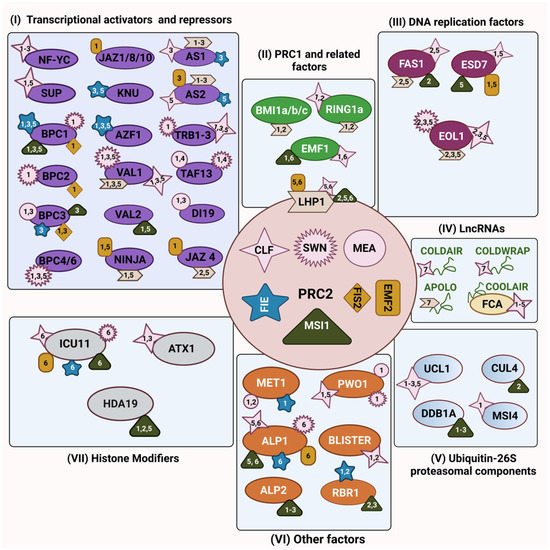
2. PRC2’s Interaction with Ubiquitin-26S Proteasomal Components
3. PRC2’s Interaction with DNA Replication Components
3.1. FASCIATA 1
3.2. ENHANCER OF LHP1 (EOL1)
3.3. DNA Polymerases
4. PRC2’s Interaction with Histone Modifiers
4.1. INCURVATA 11 (ICU11)
4.2. ARABIDOPSIS HOMOLOG OF TRITHORAX 1 (ATX1)
4.3. HISTONE DEACETYLASES (HDAC)
5. Conclusions and Perspectives
This entry is adapted from the peer-reviewed paper 10.3390/epigenomes6010008
References
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349.
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708.
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296.
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb Repressive Complex 2 in Eukaryotes—An Evolutionary Perspective. Epigenomes 2022, 6, 3.
- Baile, F.; Gómez-Zambrano, A.; Calonje, M. Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 2021, 100267.
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423.
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the gate closed: Functions of the polycomb repressive complex PRC2 in development. Plant J. 2015, 83, 121–132.
- Deevy, O.; Bracken, A.P. PRC2 functions in development and congenital disorders. Development 2019, 146, dev181354.
- Deng, W.; Buzas, D.M.; Ying, H.; Robertson, M.; Taylor, J.; Peacock, W.J.; Dennis, E.S.; Helliwell, C. Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genom. 2013, 14, 593.
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129.
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic Regulation of H3K27 Trimethylation during Arabidopsis Differentiation. PLoS Genet. 2011, 7, e1002040.
- Shu, J.; Chen, C.; Thapa, R.K.; Bian, S.; Nguyen, V.; Yu, K.; Yuan, Z.-C.; Liu, J.; Kohalmi, S.E.; Li, C.; et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 2019, 3, e00100.
- Huan, Q.; Mao, Z.; Chong, K.; Zhang, J. Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytol. 2018, 219, 1373–1387.
- Payá-Milans, M.; Poza-Viejo, L.; Martín-Uriz, P.S.; Lara-Astiaso, D.; Wilkinson, M.D.; Crevillén, P. Genome-wide analysis of the H3K27me3 epigenome and transcriptome in Brassica rapa. GigaScience 2019, 8, 1–13.
- He, G.; Zhu, X.; Elling, A.A.; Chen, L.; Wang, X.; Guo, L.; Liang, M.; He, H.; Zhang, H.; Chen, F.; et al. Global Epigenetic and Transcriptional Trends among Two Rice Subspecies and Their Reciprocal Hybrids. Plant Cell 2010, 22, 17–33.
- Makarevitch, I.; Eichten, S.; Briskine, R.; Waters, A.J.; Danilevskaya, O.N.; Meeley, R.B.; Myers, C.L.; Vaughn, M.; Springer, N.M. Genomic Distribution of Maize Facultative Heterochromatin Marked by Trimethylation of H3K27. Plant Cell 2013, 25, 780–793.
- De Lucia, F. Epigenetic Control by Plant Polycomb Proteins: New Perspectives and Emerging Roles in Stress Response; Woodhead Publishing Limited: Sawston, UK, 2013; ISBN 9781907568299.
- Shen, Q.; Lin, Y.; Li, Y.; Wang, G. Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 2021, 10, 1165.
- Mozgová, I.; Muñoz-Viana, R.; Hennig, L. PRC2 Represses Hormone-Induced Somatic Embryogenesis in Vegetative Tissue of Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006562.
- Chen, T.; Dent, S.Y.R. Chromatin modifiers: Regulators of cellular differentiation Taiping. Nat Rev Genet. 2014, 15, 93–106.
- Yang, Z.; Qian, S.; Scheid, R.N.; Lu, L.; Chen, X.; Liu, R.; Du, X.; Lv, X.; Boersma, M.D.; Scalf, M.; et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat. Genet. 2018, 50, 1247–1253.
- Li, Z.; Fu, X.; Wang, Y.; Liu, R.; He, Y. Polycomb-mediated gene silencing by the BAH–EMF1 complex in plants. Nat. Genet. 2018, 50, 1254–1261.
- Nashun, B.; Hill, P.W.S.; Hajkova, P. Reprogramming of cell fate: Epigenetic memory and the erasure of memories past. EMBO J. 2015, 34, 1296–1308.
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendía, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629.
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.; Tomlins, S.; Mehra, R.; Laxman, B.; Cao, X.; Kleer, C.G.; et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008, 27, 7274–7284.
- Casanova, M.; Preissner, T.; Cerase, A.; Poot, R.; Yamada, D.; Li, X.; Appanah, R.; Bezstarosti, K.; Demmers, J.; Koseki, H.; et al. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development 2011, 138, 1471–1482.
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281.
- Huo, Y.; Yan, Z.; Zhang, B.; Wang, X. Recruitment of Polycomb Repressive Complex 2 is Essential to Suppress the Target Chromatin in Arabidopsis. Crit. Rev. Plant Sci. 2016, 35, 131–145.
- Hepworth, J.; Dean, C. Flowering Locus C’s Lessons: Conserved Chromatin Switches Underpinning Developmental Timing and Adaptation. Plant Physiol. 2015, 168, 1237–1245.
- Xu, S.; Chong, K. Remembering winter through vernalisation. Nat. Plants 2018, 4, 997–1009.
- Sharma, N.; Geuten, K.; Giri, B.S.; Varma, A. The molecular mechanism of vernalization in Arabidopsis and cereals: Role of Flowering Locus C and its homologs. Physiol. Plant. 2020, 170, 373–383.
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-Mediated H2Aub Initiate the Switch from Embryonic to Postgerminative Growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329.
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878.
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459.
- Callis, J. The Ubiquitination Machinery of the Ubiquitin System. Arab. Book 2014, 12, e0174.
- March, E.; Farrona, S. Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones. Front. Plant Sci. 2018, 8, 2274.
- Jeong, C.W.; Roh, H.; Dang, T.V.; Choi, Y.D.; Fischer, R.L.; Lee, J.S. An E3 ligase complex regulates SET-domain polycomb group protein activity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8036–8041.
- Higa, L.A.; Zhang, H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2007, 2, 5.
- Dumbliauskas, E.; Lechner, E.; Jaciubek, M.; Berr, A.; Pazhouhandeh, M.; Alioua, M.; Cognat, V.; Brukhin, V.; Koncz, C.; Grossniklaus, U.; et al. The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 2011, 30, 731–743.
- Pazhouhandeh, M.; Molinier, J.; Berr, A.; Genschik, P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 3430–3435.
- Meinke, D.W. Genome-wide identification of EMBRYO—DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2019, 226, 306–325.
- Zhang, Y.; Li, Z.; Chen, N.; Huang, Y.; Huang, S. Phase separation of Arabidopsis emb1579 controls transcription, mRNA splicing, and development. PLoS Biol. 2020, 18, e3000782.
- Seif, E.; Kang, J.J.; Sasseville, C.; Senkovich, O.; Kaltashov, A.; Boulier, E.L.; Kapur, I.; Kim, C.A.; Francis, N.J. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat. Commun. 2020, 11, 1–19.
- Santos, A.P.; Gaudin, V.; Mozgová, I.; Pontvianne, F.; Schubert, D.; Tek, A.L.; Dvořáčková, M.; Liu, C.; Fransz, P.; Rosa, S.; et al. Tidying-up the plant nuclear space: Domains, functions, and dynamics. J. Exp. Bot. 2020, 71, 5160–5178.
- Gaillard, P.-H.; Martini, E.M.-D.; Kaufman, P.; Stillman, B.; Moustacchi, E.; Almouzni, G. Chromatin Assembly Coupled to DNA Repair: A New Role for Chromatin Assembly Factor I. Cell 1996, 86, 887–896.
- Hammond, C.; Strømme, C.B.; Huang, H.; Patel, H.H.D.J.; Groth, C.M.H.C.B.S.A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158.
- Kaya, H.; Shibahara, K.-I.; Taoka, K.-I.; Iwabuchi, M.; Stillman, B.; Araki, T. FASCIATA Genes for Chromatin Assembly Factor-1 in Arabidopsis Maintain the Cellular Organization of Apical Meristems. Cell 2001, 104, 131–142.
- Exner, V.; Taranto, P.; Schönrock, N.; Gruissem, W.; Hennig, L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 2006, 133, 4163–4172.
- Jiang, D.; Berger, F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 2017, 357, 1146–1149.
- Villa, F.; Simon, A.C.; Bazan, M.A.O.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396.
- Zhou, Y.; Tergemina, E.; Cui, H.; Förderer, A.; Hartwig, B.; James, G.V.; Schneeberger, K.; Turck, F. Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 4833–4838.
- Del Olmo, I.; López-González, L.; Martín-Trillo, M.M.; Martínez-Zapater, J.M.; Piñeiro, M.; Jarillo, J.A. Early in short days 7(ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010, 61, 623–636.
- Jenik, P.D.; Jurkuta, R.E.; Barton, M.K. Interactions between the Cell Cycle and Embryonic Patterning in Arabidopsis Uncovered by a Mutation in DNA Polymerase ε. Plant Cell 2005, 17, 3362–3377.
- Del Olmo, I.; López, J.A.; Vázquez, J.; Raynaud, C.; Piñeiro, M.; Jarillo, J.A. Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 2016, 44, 5597–5614.
- Pedroza-Garcia, J.-A.; De Veylder, L.; Raynaud, C. Plant DNA Polymerases. Int. J. Mol. Sci. 2019, 20, 4814.
- Bloomer, R.H.; Hutchison, C.E.; Bäurle, I.; Walker, J.; Fang, X.; Perera, P.; Velanis, C.N.; Gümüs, S.; Spanos, C.; Rappsilber, J.; et al. The Arabidopsis epigenetic regulator ICU11 as an accessory protein of Polycomb Repressive Complex 2. Proc. Natl. Acad. Sci. USA 2020, 117, 16660–16666.
- Dodd, I.; Micheelsen, M.A.; Sneppen, K.; Thon, G. Theoretical Analysis of Epigenetic Cell Memory by Nucleosome Modification. Cell 2007, 129, 813–822.
- Sneppen, K.; Ringrose, L. Theoretical analysis of Polycomb-Trithorax systems predicts that poised chromatin is bistable and not bivalent. Nat. Commun. 2019, 10, 1–18.
- Yang, H.; Howard, M.; Dean, C. Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. USA 2016, 113, 9369–9374.
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis Homolog of Trithorax, Activates Flower Homeotic Genes. Curr. Biol. 2003, 13, 627–637.
- Saleh, A.; Al-Abdallat, A.; Ndamukong, I.; Alvarez-Venegas, R.; Avramova, Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007, 35, 6290–6296.
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326.
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713.
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353.
- Jang, I.-C.; Chung, P.J.; Hemmes, H.; Jung, C.; Chua, N.-H. Rapid and Reversible Light-Mediated Chromatin Modifications of Arabidopsis Phytochrome A Locus. Plant Cell 2011, 23, 459–470.
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520–1523.
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2015, 28, 42–54.
- Baile, F.; Merini, W.; Hidalgo, I.; Calonje, M. EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis. Plant Cell 2021, 33, 2701–2715.
- Zeng, X.; Gao, Z.; Jiang, C.; Yang, Y.; Liu, R.; He, Y. HISTONE DEACETYLASE 9 Functions with Polycomb Silencing to Repress FLOWERING LOCUS C Expression. Plant Physiol. 2019, 182, 555–565.
- Chen, N.; Veerappan, V.; Abdelmageed, H.; Kang, M.; Allen, R.D. HSI2/VAL1 Silences AGL15 to Regulate the Developmental Transition from Seed Maturation to Vegetative Growth in Arabidopsis. Plant Cell 2018, 30, 600–619.
- Chhun, T.; Chong, S.Y.; Park, B.S.; Wong, E.C.C.; Yin, J.-L.; Kim, M.; Chua, N.-H. HSI2 Repressor Recruits MED13 and HDA6 to Down-Regulate Seed Maturation Gene Expression Directly During Arabidopsis Early Seedling Growth. Plant Cell Physiol. 2016, 57, 1689–1706.
- Zhou, S.; Liu, X.; Zhou, C.; Zhou, Q.; Zhao, Y.; Li, G.; Zhou, D.-X. Cooperation between the H3K27me3 chromatin marker and non-CG methylation in epigenetic regulation. Plant Physiol. 2016, 172, 1131–1141.
- Huang, Y.; Chen, D.-H.; Liu, B.-Y.; Shen, W.-H.; Ruan, Y. Conservation and diversification of polycomb repressive complex 2 (PRC2) proteins in the green lineage. Brief. Funct. Genom. 2016, 16, 106–119.
