Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
The proteasome system is a large and complex molecular machinery responsible for the degradation of misfolded, damaged, and redundant cellular proteins. When proteasome function is impaired, unwanted proteins accumulate, which can lead to several diseases including age-related and neurodegenerative diseases. Enhancing proteasome-mediated substrate degradation with small molecules may therefore be a valuable strategy for the treatment of various neurodegenerative diseases such as Parkinson’s, Alzheimer’s, and Huntington’s diseases.
- proteasome
- neurodegeneration
- ubiquitin
- misfolded
- disordered
- degradation
- protein
1. Introduction
The degradation of proteins is a continual process that is highly regulated by the two major proteolysis systems, the lysosomal degradation pathway and the proteasome-mediated pathway. Protein degradation helps maintain biological homeostasis in cells which are needed for all cell functions and for maintaining optimal conditions for enzyme function [1]. The proteasome pathway is the major pathway for the degradation of misfolded, oxidatively damaged, and redundant proteins. Dysregulation of proteasome function has been identified in the pathogenesis of several neurodegenerative diseases including Parkinson’s disease (PD) [2], Alzheimer’s disease (AD), and other neurodegenerative diseases [3]. The proteasome pathway is also involved in the regulation of several other cellular processes such as cell cycle, stress signaling, gene expression regulation, inflammatory response, cell differentiation, and apoptosis, which makes it an appealing target in the treatment of other types of diseases, including cancer [4]. Due to the critical role of the proteasome-mediated degradation pathway in cell regulation, the modulation of proteasome proteolytic activity has become a valuable strategy in the pursuit of new therapeutics to treat several neurodegenerative diseases [5][6][7][8].
1.1. The Human Proteasome
The human proteasome is a large complex protein responsible for the intracellular degradation of unwanted and damaged proteins via a ubiquitin-dependent and ubiquitin-independent degradation pathway. The most common proteolytic clearance of proteins proceeds by tagging the protein with polyubiquitin, after which it is degraded into small peptides of seven to eight amino acids by the 26S proteasome [9]. Highly disordered proteins can also be degraded in a ubiquitin-independent manner by the 20S proteasome [10].
1.2. Ubiquitin-Proteasome System
1.2.1. Ubiquitin
Ubiquitin (Ub) is a small protein (approximately 8600 Da) with 76 amino acid residues responsible for tagging a wide range of cellular proteins for proteolytic degradation. In the ubiquitin-proteasome system (UPS) (Figure 1), proteins are tagged for proteolysis by covalent ligation to ubiquitin [11]. Ubiquitination of proteins requires three enzymes in chronological order (see Figure 1a). The E1 ubiquitin-activating enzyme, just like its name, activates the C-terminal glycine residue of the ubiquitin in an ATP-dependent manner. The binding of the ubiquitin to a cysteine residue of E1 forms a Ub-E1 complex via a thioester linkage. The E2 ubiquitin-conjugating enzymes transfer the ubiquitin from the Ub-E1 complex to itself via a trans-thioesterification to form the Ub-E2 complex and release the E1 enzyme from the system. Lastly, the ubiquitin ligases E3s are responsible for selecting proteins for ubiquitin-mediated proteolysis. Humans have two E1 enzymes, about 40 E2 enzymes, and are estimated to have about 500–1000 E3s [12].
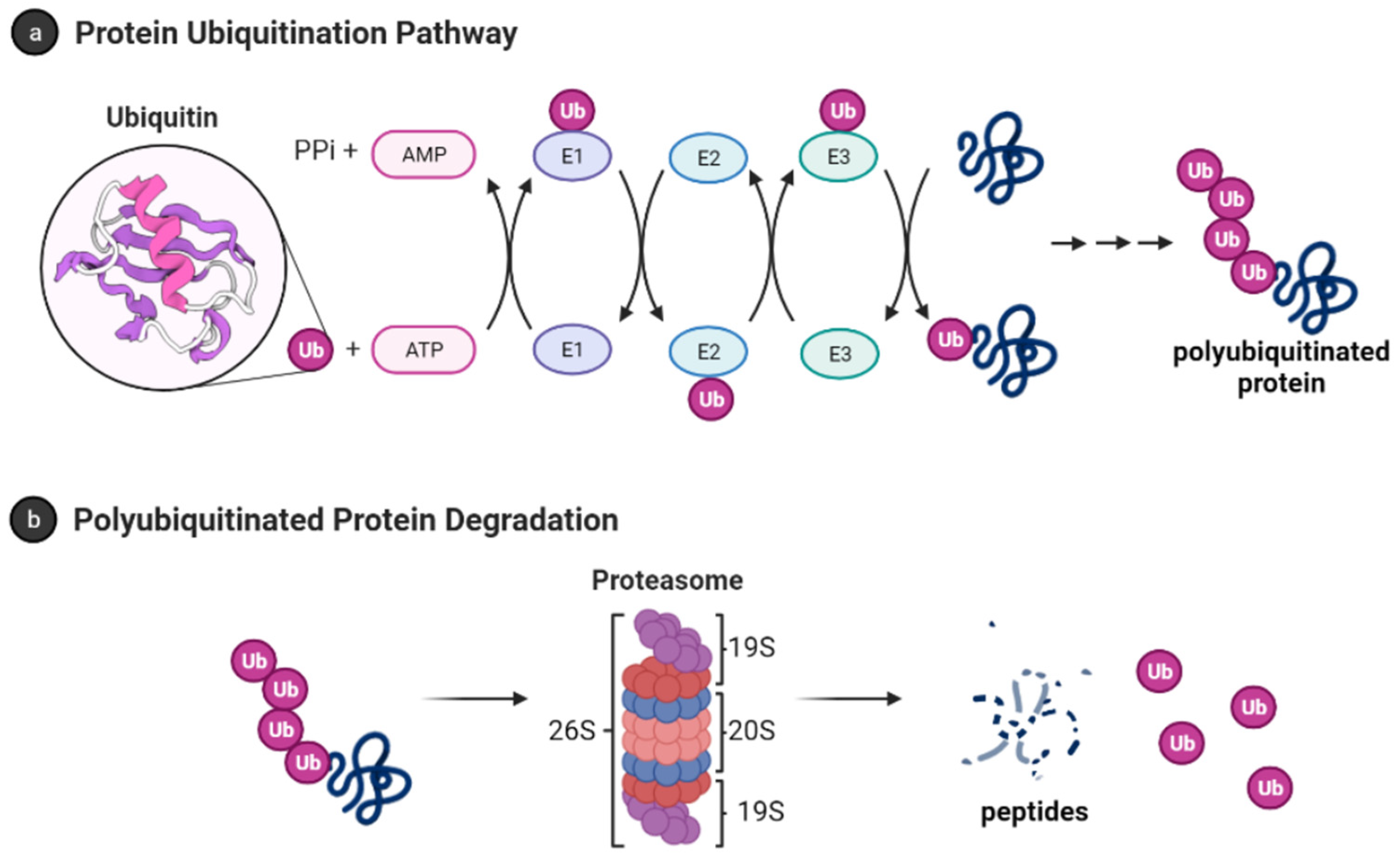
Figure 1. Ubiquitin-proteasome system [13]. (a) Protein polyubiquitination process using the ubiquitin-activating enzyme E1, conjugating enzymes E2 and the E3 ligase; (b) Polyubiquitinated proteins are degraded by 26S proteasome into small peptides following its deubiquitination.
After monoubiquitination of the targeted protein, the C-terminus of each ubiquitin molecule can be linked to any of the other seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) on the previous ubiquitin to extend the ubiquitin chain and form the polyubiquitinated tagged protein [14][15]. However, the signal for protein degradation by the proteasome usually involves the linking of Ub to the K48 of the previous Ub on the protein [16][17]. In addition, K11, K29, and K63 linked chains have also been shown to play a role in proteasomal degradation [17][18]. The 26S proteasome degrades polyubiquitinated proteins (see Figure 1b), and a previous study shows that proteins marked for degradation must be tagged with at least four ubiquitin molecules to be recognized by the 26S proteasome [16][19]. However, shorter chains, monoubiquitinated and multiple monoubiquitinated proteins can also be targeted for degradation by the proteasome [20][21][22][23]. It is also important to note that the ubiquitination process is reversible, and the deubiquitinating enzymes are present in the cell to remove ubiquitin-tagged proteins [24].
1.2.2. The 26S Proteasome
The 26S proteasome has a molecular weight of approximately 2.5 MDa and it is made up of the 20S core particle (CP), and one or two 19S regulatory particle(s) (RP) attached to one or both end(s) of the CP [25]. The 19S RP (also known as PA700) binds to the 20S CP and facilitates the gate opening of the CP for proteolytic degradation of polyubiquitinated proteins [26]. The 19S RP is also responsible for recognizing, unfolding, and translocating polyubiquitinated protein into the 20S CP [27]. Cryo-EM studies have shown many conformation states of the 26S proteasome when engaged with substrate [28][29][30][31][32][33][34][35]. Some of these studies showed the processes by which substrate is engaged, deubiquitylated, unfolded, and translocated by the proteasome [28][29]. The proteasome is also referred to as the 30S proteasome when the 20S CP is capped at both ends with the 19S RP [36]. However, we will refer to the 26S proteasome without distinguishing between the singly or doubly capped CP.
1.3. The 20S Proteasome or Core Particle
The 20S proteasome is a 700 kDa protein with a cylindrical-like structure. The CP contains four heptameric rings stacked on each other in an α1-7β1-7β1-7α1-7 fashion. The outer α-rings form a gate, and they recognize regulatory particles that allow the opening and closing of the gate [37]. The inner β-rings contains six proteolytic sites, three on each β-ring (β1, β2, and β5), and are responsible for the proteolytic activity of the proteasome.
The three different proteolytic sites of the 20S CP exhibit different substrate preferences even though they all use N-terminal nucleophilic threonine to carry out their proteolytic activities. The β1 exhibits a caspase-like (C-L)/PGPH (peptidylglutamyl-peptide hydrolyzing) activity and preferentially cleaves after acidic residues. The β2 and β5 exhibit trypsin-like (T-L) and chymotrypsin-like (CT-L) activities, and they preferentially cleave after basic and hydrophobic residues, respectively [38]. The 20S proteasome on its own degrades unstructured proteins using a ubiquitin-independent pathway.
1.4. Small Molecule Regulation of Proteasome Function
Due to the role of the proteasome in cellular functions, the regulation of proteasome has become a valuable target for the development of therapeutic molecules [39]. Proteasome inhibition is a therapeutic approach for the treatment of cancer. For example, bortezomib, a dipeptide boronate, was approved by the FDA in 2003 as an anticancer drug to treat mantle cell lymphoma and multiple myeloma [40][41]. Bortezomib inhibits the 26S proteasome by forming a covalent bond between its boron atom and threonine oxygen in the CT-L catalytic site of the 20S CP [40]. Molecules that inhibit the proteasome have also been shown to induce apoptosis in cell cultures and murine models of cancer. One of the proposed mechanisms is that proteasome inhibition prevents the degradation of the IκB, an NF-κB inhibitor, which prevents NF-κB nuclear translocation and consequently NF-κB mediated gene expression [42]. Proteasome inhibition results in the accumulation of IκB [43][44][45][46][47][48], cyclin-dependent kinase (CDK) inhibitor p21 [43][49][50], tumor suppressor p53, and other pro-apoptotic proteins [51][52][53]. The exceptional increase in apoptosis of certain multiple myeloma cells when treated with proteasome inhibitors has also been linked to an increase in protein unfolding and increasing substrate load on the proteasomes [54][55]. In addition, proteasome inhibition leads to lethal shortage of amino acids in the cells, which are the building blocks for cells to make new proteins. This amino acid scarcity caused by proteasome inhibition results in increasing ER stress and cell apoptosis [56]. Many reviews on proteasome inhibition have recently been published [57][58][59][60][61][62][63][64][65][66], including a recent review by our group on natural products scaffolds as inhibitors of the proteasome [67].
Proteasome activation by small molecules is a proposed strategy for the treatment of age-related diseases and several neurodegenerative diseases such as Parkinson’s disease (PD), Alzheimer’s Disease (AD), Huntington’s Disease (HD), and Amyotrophic Lateral Sclerosis [8][68][69][70][71][72]. Increasing the proteolytic activity of proteasome enhances the degradation of specific intrinsically disordered proteins (IDPs) such as α-synuclein, β-amyloid, and tau, to mention a few, which are associated with the pathogenesis of these neurodegenerative diseases.
2. Proteasome Activity and Diseases
As humans age, there is a decline in proteasome function [3][73]. This reduction could be due to the reduction in the expression of proteasome subunits [74], oxidative damage of the protein [75][76], and disassembly of the 26S proteasome holocomplex [77][78]. The decrease in proteasome proteolytic function leads to lower rates of unwanted protein degradation which can induce toxic signaling upon accumulation and aggregation (Figure 2). In particular, the accumulation of specific intrinsically disordered proteins (IDPs), such as amyloid-β and α-synuclein, have been identified as a driving cause of many neurodegenerative diseases [79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98]. The exact mechanism by which these oligomers induce neurotoxicity is complex and still debated, but it is widely accepted that dysregulated IDPs accumulate, and the resulting soluble oligomeric forms of these protein aggregates are likely toxic species in disease pathogenesis [79][93][99][100][101][102]. These soluble oligomeric forms are also responsible for impairing proteasome function, which further drives disease progression [94][103][104][105][106][107][108][109][110][111][112][113][114][115][116][117][118][119][120]. Multiple studies have indicated that enhancing proteasome proteolytic activity prevents the accumulation of these IDPs, reduces brain damage and improves cognitive performance in mouse models, and may be a new therapeutic strategy to treat neurodegenerative diseases [8][68][69][70][72][119][121][122][123][124][125][126][127][128][129][130][131][132]. More recently, it has been recognized that the 20S proteasome of the proteasome plays a critical role in maintaining proteostasis by the direct degradation of oxidatively damaged and highly disordered proteins [10][133][134][135][136][137][138]. The 20S proteasome, therefore, serves as the default protease to unremittently maintain low levels of these unwanted IDPs without the need for post-translation modifications, including protein ubiquitination [10][133]. Highly disordered proteins appear to be the main target of the 20S proteasome [139]. IDPs are also naturally short-lived, but basal levels are secured by forming proteolytically stable structured complexes with “nannies”, chaperones, or other protein complexes [140]. However, when these IDPs production outpaces their degradation, they accumulate, oligomerize, and aggregate, resulting in the induction of downstream cytotoxic signaling events.
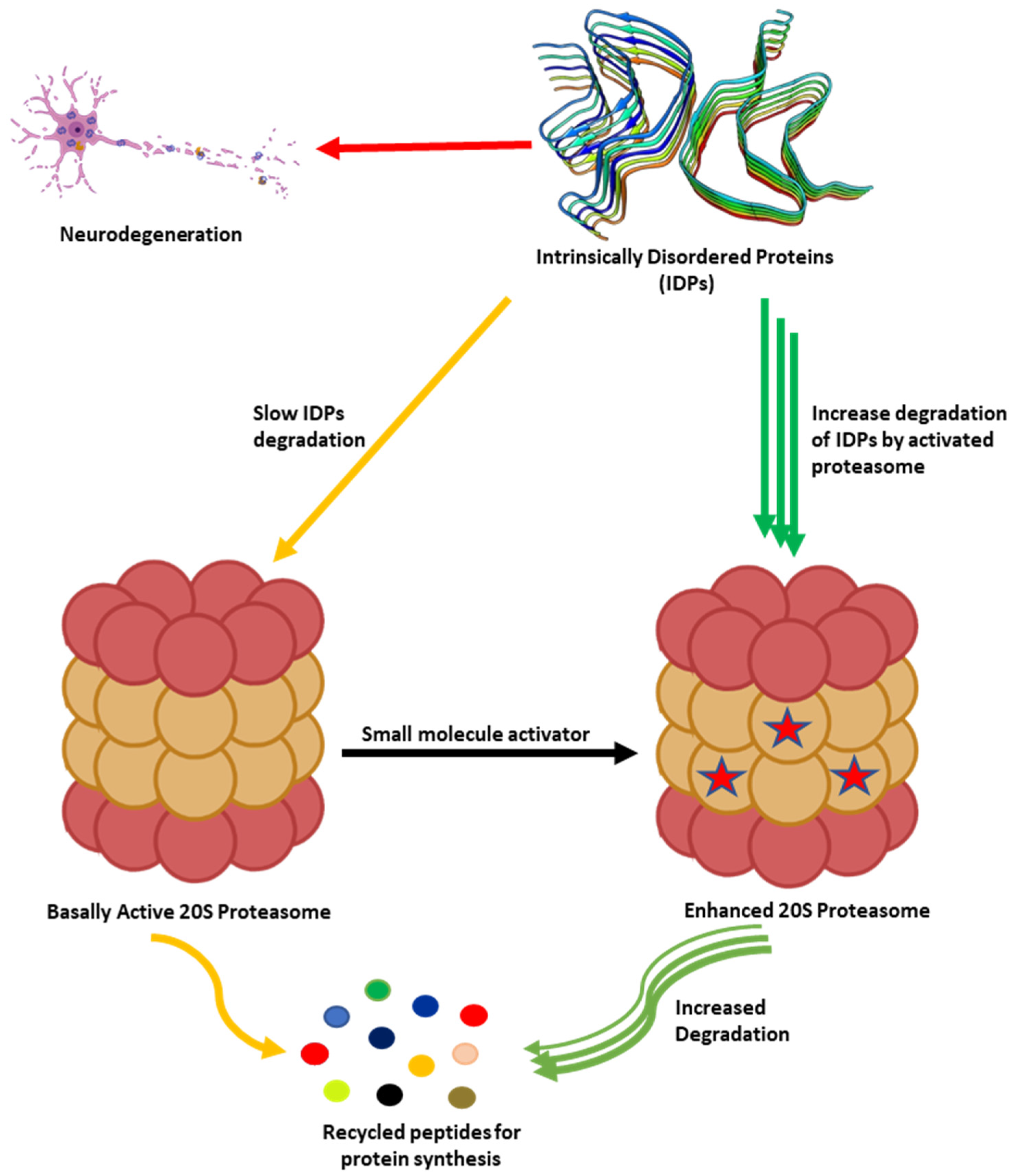
Figure 2. Accumulation of partially unfolded, misfolded, and dysregulated intrinsically disordered proteins (IDPs) such as amyloid-β and α-synuclein leads to neurotoxicity and neuronal cell death. The 20S proteasome degrades unwanted IDPs; however, small molecules can enhance the rate of proteasome-mediated degradation of these IDPs and prevent their accumulation.
3. Conclusions
Efficient proteasome function is critical in maintaining healthy cellular homeostasis. Dysregulation of protein or proteasome impairment can result in a toxic accumulation of unwanted proteins, which is observed in the pathogenesis of different neurodegenerative diseases and aging. Enhancing the proteolytic activity of the proteasome by increasing its capacity, accessibility, or the rate at which it degrades has long been hypothesized as a means to prevent the accumulation of dysregulated IDPs. More recently, researchers from various labs have explored the use of small molecules to induce protein proteolysis. Small molecule proteasome agonists can enhance the proteolytic clearance of unwanted proteins and restore homeostasis. Small molecule enhancers of the 26S proteasome mainly induce enhanced 26S-mediated proteolysis of ubiquitinated proteins via an indirect mechanism of proteasome activation.
Small molecule inhibitors of deubiquitinases prevent proteins marked for ubiquitin-dependent degradation from escaping their fate. Even though there are no approved therapies yet based on deubiquitinating enzyme (DUB) inhibitors, this is an emerging field with great significance. Small molecule regulation of upstream signaling pathways, including cAMP-depending protein kinase A and c-GMP-dependent protein kinase G, affect the phosphorylation of the proteasome regulatory particles. As a result, small molecule regulators of phosphodiesterase type-3 (PDE3) can therefore indirectly increase the rate of substrate degradation by the proteasome. Small molecules that directly interact with the 26S proteasome and enhance the rate of 26S proteasome-mediated protein degradation are less known and likely a fruitful field for exploration.
Whereas the 26S proteasome targets ubiquitinylated protein substrates, the 20S proteasome is limited to the degradation of only disordered proteins. Several small molecule enhancers of 20S proteasome-mediated protein degradation have been identified in the literature that induce 20S—mediated degradation of dysregulated intrinsically disordered proteins by direct interaction with the 20S core particle.
The activation of the proteasome by small molecules is a relatively new field in science. Its potential as a therapeutic approach is still unknown and the consequences of chronic exposure to proteasome enhancers are not known. However, considering the possibility of treating multiple disorders for which there are currently no treatment options available, this approach has enormous potential. However, as in all new fields, the approach still needs further validation, in vivo studies in particular, to fully understand its therapeutic potential and limitation. In addition, more studies are needed to elucidate the mechanistic details of small molecule proteasome activation and its overall cellular consequences.
This entry is adapted from the peer-reviewed paper 10.3390/biom11121789
References
- Hetz, C.; Glimcher, L.H. Protein homeostasis networks in physiology and disease. Curr. Opin. Cell. Biol. 2011, 23, 123–125.
- McNaught, K.S.P.; Olanow, C.W.; Halliwell, B.; Isacson, O.; Jenner, P. Failure of the ubiquitin–proteasome system in Parkinson’s disease. Nat. Rev. Neurosci. 2001, 2, 589–594.
- Saez, I.; Vilchez, D. The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases. Curr. Genomics 2014, 15, 38–51.
- LaPlante, G.; Zhang, W. Targeting the Ubiquitin-Proteasome System for Cancer Therapeutics by Small-Molecule Inhibitors. Cancers 2021, 13, 3079.
- Momtaz, S.; Memariani, Z.; El-Senduny, F.F.; Sanadgol, N.; Golab, F.; Katebi, M.; Abdolghaffari, A.H.; Farzaei, M.H.; Abdollahi, M. Targeting Ubiquitin-Proteasome Pathway by Natural Products: Novel Therapeutic Strategy for Treatment of Neurodegenerative Diseases. Front. Physiol. 2020, 11, 361.
- Rao, G.; Croft, B.; Teng, C.; Awasthi, V. Ubiquitin-Proteasome System in Neurodegenerative Disorders. J. Drug Metab. Toxicol. 2015, 6, 187.
- Huang, Q.; Figueiredo-Pereira, M.E. Ubiquitin/proteasome pathway impairment in neurodegeneration: Therapeutic implications. Apoptosis 2010, 15, 1292–1311.
- Njomen, E.; Tepe, J.J. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J. Med. Chem. 2019, 62, 6469–6481.
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371.
- Kumar Deshmukh, F.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190.
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479.
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440.
- Adapted from “Ubiquitin Proteasome System”, by BioRender.com. Available online: https://app.biorender.com/biorender-templates (accessed on 19 October 2021).
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953.
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926.
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583.
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1.
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem 2012, 81, 203–229.
- hrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102.
- Dimova, N.V.; Hathaway, N.A.; Lee, B.-H.; Kirkpatrick, D.S.; Berkowitz, M.L.; Gygi, S.P.; Finley, D.; King, R.W. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 2012, 14, 168–176.
- Shabek, N.; Herman-Bachinsky, Y.; Buchsbaum, S.; Lewinson, O.; Haj-Yahya, M.; Hejjaoui, M.; Lashuel, H.A.; Sommer, T.; Brik, A.; Ciechanover, A. The Size of the Proteasomal Substrate Determines Whether Its Degradation Will Be Mediated by Mono- or Polyubiquitylation. Mol. Cell 2012, 48, 87–97.
- Kravtsova-Ivantsiv, Y.; Ciechanover, A. Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 2012, 125, 539–548.
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477.
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563.
- Peters, J.M.; Cejka, Z.; Harris, J.R.; Kleinschmidt, J.A.; Baumeister, W. Structural features of the 26S proteasome complex. J. Mol. Biol. 1993, 234, 932–937.
- da Fonseca, P.C.; Morris, E.P. Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 2008, 283, 23305–23314.
- Orlowski, M.; Wilk, S. Catalytic Activities of the 20S Proteasome, a Multicatalytic Proteinase Complex. Arch. Biochem. Biophys. 2000, 383, 1–16.
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55.
- Finley, D.; Chen, X.; Walters, K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci 2016, 41, 77–93.
- Andres, H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science 2018, 362, eaav0725.
- Eisele, M.R.; Reed, R.G.; Rudack, T.; Schweitzer, A.; Beck, F.; Nagy, I.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Tomko, R.J.; et al. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep. 2018, 24, 1301–1315.e1305.
- Ding, Z.; Fu, Z.; Xu, C.; Wang, Y.; Wang, Y.; Li, J.; Kong, L.; Chen, J.; Li, N.; Zhang, R.; et al. High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res. 2017, 27, 373–385.
- Huang, X.; Luan, B.; Wu, J.; Shi, Y. An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 2016, 23, 778–785.
- Chen, S.; Wu, J.; Lu, Y.; Ma, Y.-B.; Lee, B.-H.; Yu, Z.; Ouyang, Q.; Finley, D.J.; Kirschner, M.W.; Mao, Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, 12991.
- Ding, Z.; Xu, C.; Sahu, I.; Wang, Y.; Fu, Z.; Huang, M.; Wong, C.C.L.; Glickman, M.H.; Cong, Y. Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations. Mol. Cell 2019, 73, 1150–1161.e1156.
- Vilchez, D.; Saez, I.; Dillin, A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 2014, 5, 5659.
- Bochtler, M.; Ditzel, L.; Groll, M.; Hartmann, C.; Huber, R. The proteasome. Annu Rev Biophys Biomol Struct 1999, 28, 295–317.
- Huber, E.M.; Heinemeyer, W.; Li, X.; Arendt, C.S.; Hochstrasser, M.; Groll, M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat. Commun. 2016, 7, 10900.
- Huang, L.; Chen, C.H. Proteasome regulators: Activators and inhibitors. Curr. Med. Chem. 2009, 16, 931–939.
- Kane, R.C.; Bross, P.F.; Farrell, A.T.; Pazdur, R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 2003, 8, 508–513.
- Bruna, J.; Udina, E.; Alé, A.; Vilches, J.J.; Vynckier, A.; Monbaliu, J.; Silverman, L.; Navarro, X. Neurophysiological, histological and immunohistochemical characterization of bortezomib-induced neuropathy in mice. Exp. Neurol. 2010, 223, 599–608.
- Gilmore, T.D.; Herscovitch, M. Inhibitors of NF-κB signaling: 785 and counting. Oncogene 2006, 25, 6887–6899.
- Hideshima, T.; Richardson, P.; Chauhan, D.; Palombella, V.J.; Elliott, P.J.; Adams, J.; Anderson, K.C. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001, 61, 3071–3076.
- Russo, S.M.; Tepper, J.E.; Baldwin, A.S., Jr.; Liu, R.; Adams, J.; Elliott, P.; Cusack, J.C., Jr. Enhancement of radiosensitivity by proteasome inhibition: Implications for a role of NF-kappaB. Int. J. Radiat. Oncol. Biol. Phys. 2001, 50, 183–193.
- Sunwoo, J.B.; Chen, Z.; Dong, G.; Yeh, N.; Crowl Bancroft, C.; Sausville, E.; Adams, J.; Elliott, P.; Van Waes, C. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin. Cancer Res. 2001, 7, 1419–1428.
- Hideshima, T.; Chauhan, D.; Richardson, P.; Mitsiades, C.; Mitsiades, N.; Hayashi, T.; Munshi, N.; Dang, L.; Castro, A.; Palombella, V.; et al. NF-kappa B as a therapeutic target in multiple myeloma. J. Biol. Chem. 2002, 277, 16639–16647.
- Tan, C.; Waldmann, T.A. Proteasome inhibitor PS-341, a potential therapeutic agent for adult T-cell leukemia. Cancer Res. 2002, 62, 1083–1086.
- Ma, M.H.; Yang, H.H.; Parker, K.; Manyak, S.; Friedman, J.M.; Altamirano, C.; Wu, Z.Q.; Borad, M.J.; Frantzen, M.; Roussos, E.; et al. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clin. Cancer Res. 2003, 9, 1136–1144.
- Shah, S.A.; Potter, M.W.; McDade, T.P.; Ricciardi, R.; Perugini, R.A.; Elliott, P.J.; Adams, J.; Callery, M.P. 26S proteasome inhibition induces apoptosis and limits growth of human pancreatic cancer. J. Cell. Biochem. 2001, 82, 110–122.
- Yang, Y.; Ikezoe, T.; Saito, T.; Kobayashi, M.; Koeffler, H.P.; Taguchi, H. Proteasome inhibitor PS-341 induces growth arrest and apoptosis of non-small cell lung cancer cells via the JNK/c-Jun/AP-1 signaling. Cancer Sci. 2004, 95, 176–180.
- Williams, S.A.; McConkey, D.J. The proteasome inhibitor bortezomib stabilizes a novel active form of p53 in human LNCaP-Pro5 prostate cancer cells. Cancer Res. 2003, 63, 7338–7344.
- Li, B.; Dou, Q.P. Bax degradation by the ubiquitin/proteasome-dependent pathway: Involvement in tumor survival and progression. Proc. Natl. Acad. Sci. USA 2000, 97, 3850–3855.
- Breitschopf, K.; Zeiher, A.M.; Dimmeler, S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J. Biol. Chem. 2000, 275, 21648–21652.
- Bianchi, G.; Oliva, L.; Cascio, P.; Pengo, N.; Fontana, F.; Cerruti, F.; Orsi, A.; Pasqualetto, E.; Mezghrani, A.; Calbi, V.; et al. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood 2009, 113, 3040–3049.
- Sha, Z.; Goldberg, A.L. Multiple myeloma cells are exceptionally sensitive to heat shock, which overwhelms their proteostasis network and induces apoptosis. Proc. Natl. Acad. Sci. USA 2020, 117, 21588.
- Suraweera, A.; Münch, C.; Hanssum, A.; Bertolotti, A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 2012, 48, 242–253.
- Aliabadi, F.; Sohrabi, B.; Mostafavi, E.; Pazoki-Toroudi, H.; Webster, T.J. Ubiquitin–proteasome system and the role of its inhibitors in cancer therapy. Open Biol. 2021, 11, 200390.
- Fricker, L.D. Proteasome Inhibitor Drugs. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 457–476.
- Gandolfi, S.; Laubach, J.P.; Hideshima, T.; Chauhan, D.; Anderson, K.C.; Richardson, P.G. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 2017, 36, 561–584.
- Ito, S. Proteasome Inhibitors for the Treatment of Multiple Myeloma. Cancers 2020, 12, 265.
- Kaplan, G.S.; Torcun, C.C.; Grune, T.; Ozer, N.K.; Karademir, B. Proteasome inhibitors in cancer therapy: Treatment regimen and peripheral neuropathy as a side effect. Free Radic. Biol. Med. 2017, 103, 1–13.
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433.
- Narayanan, S.; Cai, C.-Y.; Assaraf, Y.G.; Guo, H.-Q.; Cui, Q.; Wei, L.; Huang, J.-J.; Ashby, C.R.; Chen, Z.-S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updates 2020, 48, 100663.
- Roeten, M.S.F.; Cloos, J.; Jansen, G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother. Pharmacol. 2018, 81, 227–243.
- Sherman, D.J.; Li, J. Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease. Molecules 2020, 25, 671.
- Zhang, X.; Linder, S.; Bazzaro, M. Drug Development Targeting the Ubiquitin–Proteasome System (UPS) for the Treatment of Human Cancers. Cancers 2020, 12, 902.
- Hubbell, G.E.; Tepe, J.J. Natural product scaffolds as inspiration for the design and synthesis of 20S human proteasome inhibitors. RSC Chem. Biol. 2020, 1, 305–332.
- Jones, C.L.; Tepe, J.J. Proteasome Activation to Combat Proteotoxicity. Molecules 2019, 24, 2841.
- Jones, C.L.; Njomen, E.; Sjogren, B.; Dexheimer, T.S.; Tepe, J.J. Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chem. Biol. 2017, 12, 2240–2247.
- Njomen, E.; Osmulski, P.A.; Jones, C.L.; Gaczynska, M.; Tepe, J.J. Small Molecule Modulation of Proteasome Assembly. Biochemistry 2018, 57, 4214–4224.
- Trippier, P.C.; Zhao, K.T.; Fox, S.G.; Schiefer, I.T.; Benmohamed, R.; Moran, J.; Kirsch, D.R.; Morimoto, R.I.; Silverman, R.B. Proteasome Activation is a Mechanism for Pyrazolone Small Molecules Displaying Therapeutic Potential in Amyotrophic Lateral Sclerosis. ACS Chem. Neurosci. 2014, 5, 823–829.
- Trader, D.J.; Simanski, S.; Dickson, P.; Kodadek, T. Establishment of a suite of assays that support the discovery of proteasome stimulators. Biochim. Biophys. Acta 2017, 1861, 892–899.
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217.
- Lee, C.-K.; Klopp, R.G.; Weindruch, R.; Prolla, T.A. Gene Expression Profile of Aging and Its Retardation by Caloric Restriction. Science 1999, 285, 1390–1393.
- Bulteau, A.-L.; Lundberg, K.C.; Humphries, K.M.; Sadek, H.A.; Szweda, P.A.; Friguet, B.; Szweda, L.I. Oxidative Modification and Inactivation of the Proteasome during Coronary Occlusion/Reperfusion*. J. Biol. Chem. 2001, 276, 30057–30063.
- Wang, X.; Yen, J.; Kaiser, P.; Huang, L. Regulation of the 26S Proteasome Complex During Oxidative Stress. Sci. Signal. 2010, 3, ra88.
- Tonoki, A.; Kuranaga, E.; Tomioka, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Miura, M. Genetic Evidence Linking Age-Dependent Attenuation of the 26S Proteasome with the Aging Process. Mol. Cell. Biol. 2009, 29, 1095–1106.
- Bajorek, M.; Finley, D.; Glickman, M.H. Proteasome Disassembly and Downregulation Is Correlated with Viability during Stationary Phase. Curr. Biol. 2003, 13, 1140–1144.
- Kayed, R.; Dettmer, U.; Lesné, S.E. Soluble endogenous oligomeric α-synuclein species in neurodegenerative diseases: Expression, spreading, and cross-talk. J. Parkinsons Dis. 2020, 10, 791–818.
- Ono, K. Alzheimer’s disease as oligomeropathy. Neurochem. Int. 2018, 119, 57–70.
- Mroczko, B.; Groblewska, M.; Litman-Zawadzka, A.; Kornhuber, J.; Lewczuk, P. Amyloid β oligomers (AβOs) in Alzheimer’s disease. J. Neural Transm. 2018, 125, 177–191.
- Gulisano, W.; Maugeri, D.; Baltrons, M.A.; Fà, M.; Amato, A.; Palmeri, A.; D’Adamio, L.; Grassi, C.; Devanand, D.; Honig, L.S. Role of amyloid-β and tau proteins in Alzheimer’s disease: Confuting the amyloid cascade. J. Alzheimer’s Dis. 2018, 64, S611–S631.
- Ghag, G.; Bhatt, N.; Cantu, D.V.; Guerrero-Munoz, M.J.; Ellsworth, A.; Sengupta, U.; Kayed, R. Soluble tau aggregates, not large fibrils, are the toxic species that display seeding and cross-seeding behavior. Protein Sci. 2018, 27, 1901–1909.
- Forloni, G.; Balducci, C. Alzheimer’s disease, oligomers, and inflammation. J. Alzheimer’s Dis. 2018, 62, 1261–1276.
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The amyloid-β oligomer hypothesis: Beginning of the third decade. J. Alzheimer’s Dis. 2018, 64, S567–S610.
- Choi, M.L.; Gandhi, S. Crucial role of protein oligomerization in the pathogenesis of Alzheimer’s and Parkinson’s diseases. FEBS J. 2018, 285, 3631–3644.
- Castillo-Carranza, D.L.; Guerrero-Muñoz, M.J.; Sengupta, U.; Gerson, J.E.; Kayed, R. α-Synuclein oligomers induce a unique toxic tau strain. Biol. Psychiatry 2018, 84, 499–508.
- Shafiei, S.S.; Guerrero-Muñoz, M.J.; Castillo-Carranza, D.L. Tau oligomers: Cytotoxicity, propagation, and mitochondrial damage. Front. Aging Neurosci. 2017, 9, 83.
- Sengupta, U.; Nilson, A.N.; Kayed, R. The role of amyloid-β oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 2016, 6, 42–49.
- Ingelsson, M. Alpha-synuclein oligomers—neurotoxic molecules in Parkinson’s disease and other Lewy body disorders. Front. Neurosci. 2016, 10, 408.
- Caárdenas-Aguayo, M.a.d.C.; Goόmez-Virgilio, L.; DeRosa, S.; Meraz-Ríos, M.A. The role of tau oligomers in the onset of Alzheimer’s disease neuropathology. ACS Chem. Neurosci. 2014, 5, 1178–1191.
- Katzmarski, N.; Ziegler-Waldkirch, S.; Scheffler, N.; Witt, C.; Abou-Ajram, C.; Nuscher, B.; Prinz, M.; Haass, C.; Meyer-Luehmann, M. Aβ oligomers trigger and accelerate Aβ seeding. Brain Pathol. 2020, 30, 36–45.
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112.
- Smith, D.M. Could a common mechanism of protein degradation impairment underlie many neurodegenerative diseases? J. Exp. Neurosci. 2018, 12, 1179069518794675.
- Gerson, J.E.; Farmer, K.M.; Henson, N.; Castillo-Carranza, D.L.; Murillo, M.C.; Sengupta, U.; Barrett, A.; Kayed, R. Tau oligomers mediate α-synuclein toxicity and can be targeted by immunotherapy. Mol. Neurodegener. 2018, 13, 1–14.
- Gerson, J.E.; Sengupta, U.; Kayed, R. Tau oligomers as pathogenic seeds: Preparation and propagation in vitro and in vivo. In Tau Protein; Humana Press: New York, NY, USA, 2017; pp. 141–157.
- Bengoa-Vergniory, N.; Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Alpha-synuclein oligomers: A new hope. Acta Neuropathol. 2017, 134, 819–838.
- Gerson, J.E.; Mudher, A.; Kayed, R. Potential mechanisms and implications for the formation of tau oligomeric strains. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 482–496.
- Brettschneider, J.; Del Tredici, K.; Lee, V.M.; Trojanowski, J.Q. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 2015, 16, 109–120.
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786.
- Selkoe, D.J. Folding proteins in fatal ways. Nature 2003, 426, 900–904.
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608.
- Cecarini, V.; Bonfili, L.; Amici, M.; Angeletti, M.; Keller, J.N.; Eleuteri, A.M. Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res. 2008, 1209, 8–18.
- Díaz-Hernández, M.; Valera, A.G.; Morán, M.A.; Gómez-Ramos, P.; Alvarez-Castelao, B.; Castaño, J.G.; Hernández, F.; Lucas, J.J. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J. Neurochem. 2006, 98, 1585–1596.
- Gregori, L.; Fuchs, C.; Figueiredo-Pereira, M.E.; Van Nostrand, W.E.; Goldgaber, D. Amyloid β-Protein Inhibits Ubiquitin-dependent Protein Degradation in Vitro (∗). J. Biol. Chem. 1995, 270, 19702–19708.
- Lindersson, E.; Beedholm, R.; Højrup, P.; Moos, T.; Gai, W.; Hendil, K.B.; Jensen, P.H. Proteasomal inhibition by α-synuclein filaments and oligomers. J. Biol. Chem. 2004, 279, 12924–12934.
- Bence, N.F.; Sampat, R.M.; Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001, 292, 1552–1555.
- Oh, S.; Hong, H.S.; Hwang, E.; Sim, H.J.; Lee, W.; Shin, S.J.; Mook-Jung, I. Amyloid peptide attenuates the proteasome activity in neuronal cells. Mech. Ageing Dev. 2005, 126, 1292–1299.
- Tanaka, K.; Matsuda, N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta 2014, 1843, 197–204.
- Tanaka, Y.; Engelender, S.; Igarashi, S.; Rao, R.K.; Wanner, T.; Tanzi, R.E.; Sawa, A.; Dawson, V.L.; Dawson, T.M.; Ross, C.A. Inducible expression of mutant α-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum. Mol. Genet. 2001, 10, 919–926.
- Tseng, B.P.; Green, K.N.; Chan, J.L.; Blurton-Jones, M.; LaFerla, F.M. Aβ inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol. Aging 2008, 29, 1607–1618.
- Emmanouilidou, E.; Stefanis, L.; Vekrellis, K. Cell-produced α-synuclein oligomers are targeted to, and impair, the 26S proteasome. Neurobiol. Aging 2010, 31, 953–968.
- Deriziotis, P.; André, R.; Smith, D.M.; Goold, R.; Kinghorn, K.J.; Kristiansen, M.; Nathan, J.A.; Rosenzweig, R.; Krutauz, D.; Glickman, M.H. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011, 30, 3065–3077.
- Deriziotis, P.; Tabrizi, S.J. Prions and the proteasome. Biochim. Biophys. Acta 2008, 1782, 713–722.
- Kristiansen, M.; Deriziotis, P.; Dimcheff, D.E.; Jackson, G.S.; Ovaa, H.; Naumann, H.; Clarke, A.R.; van Leeuwen, F.W.; Menéndez-Benito, V.; Dantuma, N.P. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol. Cell 2007, 26, 175–188.
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097.
- Zondler, L.; Kostka, M.; Garidel, P.; Heinzelmann, U.; Hengerer, B.; Mayer, B.; Weishaupt, J.H.; Gillardon, F.; Danzer, K.M. Proteasome impairment by α-synuclein. PLoS ONE 2017, 12, e0184040.
- Ruegsegger, C.; Saxena, S. Proteostasis impairment in ALS. Brain Res. 2016, 1648, 571–579.
- Myeku, N.; Clelland, C.L.; Emrani, S.; Kukushkin, N.V.; Yu, W.H.; Goldberg, A.L.; Duff, K.E. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 2016, 22, 46–53.
- Papanikolopoulou, K.; Skoulakis, E. Altered proteostasis in neurodegenerative tauopathies. In Proteostasis and Disease; Barrio, R., Sutherland, J.D., Rodriguez, M.S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 177–194.
- Choi, W.H.; De Poot, S.A.; Lee, J.H.; Kim, J.H.; Han, D.H.; Kim, Y.K.; Finley, D.; Lee, M.J. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat. Commun. 2016, 7, 1–12.
- Leestemaker, Y.; de Jong, A.; Witting, K.F.; Penning, R.; Schuurman, K.; Rodenko, B.; Zaal, E.A.; van de Kooij, B.; Laufer, S.; Heck, A.J. Proteasome activation by small molecules. Cell Chem. Biol. 2017, 24, 725–736.
- Leestemaker, Y.; Ovaa, H. Tools to investigate the ubiquitin proteasome system. Drug Discov. Today Technol. 2017, 26, 25–31.
- Lee, B.-H.; Lee, M.J.; Park, S.; Oh, D.-C.; Elsasser, S.; Chen, P.-C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184.
- Fiolek, T.J.; Magyar, C.L.; Wall, T.J.; Davies, S.B.; Campbell, M.V.; Savich, C.J.; Tepe, J.J.; Mosey, R.A. Dihydroquinazolines enhance 20S proteasome activity and induce degradation of alpha-synuclein, an intrinsically disordered protein associated with neurodegeneration. Bioorg. Med. Chem. Lett. 2021, 36, 127821.
- Coleman, R.A.; Muli, C.S.; Zhao, Y.; Bhardwaj, A.; Newhouse, T.R.; Trader, D.J. Analysis of chain length, substitution patterns, and unsaturation of AM-404 derivatives as 20S proteasome stimulators. Bioorg. Med. Chem. Lett. 2019, 29, 420–423.
- Coleman, R.A.; Trader, D.J. Development and Application of a Sensitive Peptide Reporter to Discover 20S Proteasome Stimulators. ACS Comb. Sci. 2018, 20, 269–276.
- Giżynńska, M.; Witkowska, J.; Karpowicz, P.; Rostankowski, R.; Chocron, E.S.; Pickering, A.M.; Osmulski, P.; Gaczynska, M.; Jankowska, E.b. Proline-and arginine-rich peptides as flexible allosteric modulators of human proteasome activity. J. Med. Chem. 2019, 62, 359–370.
- Njomen, E.; Lansdell, T.; Vanecek, A.; Benham, V.; Bernard, M.; Yang, Y.-T.; Schall, P.; Isaac, D.; Alkharabsheh, O.; Al-Janadi, A. Enhancing c-MYC degradation via 20S proteasome activation induces in vivo anti-tumor efficacy. bioRxiv 2020.
- Chondrogianni, N.; Georgila, K.; Kourtis, N.; Tavernarakis, N.; Gonos, E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J. 2015, 29, 611–622.
- Gonos, E. Proteasome activation as a novel anti-aging strategy. Free Radic. Biol. Med. 2014, 75, S7.
- Fiolek, T.J.; Keel, K.L.; Tepe, J.J. Fluspirilene Analogs Activate the 20S Proteasome and Overcome Proteasome Impairment by Intrinsically Disordered Protein Oligomers. ACS Chem. Neurosci. 2021, 12, 1438–1448.
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884.
- Njomen, E.; Tepe, J.J. Regulation of Autophagic Flux by the 20S Proteasome. Cell Chem. Biol. 2019, 26, 1283–1294.
- Asher, G.; Reuven, N.; Shaul, Y. 20S proteasomes and protein degradation "by default". Bioessays 2006, 28, 844–849.
- Korovila, I.; Hugo, M.; Castro, J.P.; Weber, D.; Höhn, A.; Grune, T.; Jung, T. Proteostasis, oxidative stress and aging. Redox Biol. 2017, 13, 550–567.
- Höhn, T.J.; Grune, T. The proteasome and the degradation of oxidized proteins: Part III-Redox regulation of the proteasomal system. Redox Biol. 2014, 2, 388–394.
- Chondrogianni, N.; Petropoulos, I.; Grimm, S.; Georgila, K.; Catalgol, B.; Friguet, B.; Grune, T.; Gonos, E.S. Protein damage, repair and proteolysis. Mol. Aspects Med. 2014, 35, 1–71.
- Myers, N.; Olender, T.; Savidor, A.; Levin, Y.; Reuven, N.; Shaul, Y. The Disordered Landscape of the 20S Proteasome Substrates Reveals Tight Association with Phase Separated Granules. Proteomics 2018, 18, e1800076.
- Tsvetkov, P.; Reuven, N.; Shaul, Y. The nanny model for IDPs. Nat. Chem. Biol. 2009, 5, 778–781.
This entry is offline, you can click here to edit this entry!
