Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Oncology
Lung cancer is the leading cause of malignancy-related mortality worldwide. AI has the potential to help to treat lung cancer from detection, diagnosis and decision making to prognosis prediction. AI could reduce the labor work of LDCT, CXR, and pathology slides reading. AI as a second reader in LDCT and CXR reading reduces the effort of radiologists and increases the accuracy of nodule detection.
- artificial intelligence
- machine learning
- lung cancer
1. Introduction
Lung cancer constitutes the largest portion of malignancy-related deaths worldwide [1]. It is also the leading cause of malignancy-related death in Taiwan [2][3]. The majority of the patients diagnosed with lung cancer are in the late-stage, and therefore have a poor prognosis. In addition to the late stage at diagnosis, the heterogeneity of imaging features and histopathology of lung cancer also makes it a challenge for clinicians to choose the best treatment option.
The imaging features of lung cancer vary from a single tiny nodule to ground-glass opacity, multiple nodules, pleural effusion, lung collapse, and multiple opacities [4]; simple and small lesions are extremely difficult to detect [5]. Histopathological features include adenocarcinoma, squamous cell carcinoma, small cell carcinoma, and many other rare histological types. The histology subtypes vary even more. For example, at least six common subtypes and a total of eleven subtypes of adenocarcinoma were reported in the 2015 World Health Organization classification of lung tumors [6], with more subtypes added to the 2021 version [7]. Treatment options are heavily dependent on the clinical staging, histopathology, and genomic features of the lung cancer. In the era of precision medicine, clinicians need to collect all the features and make a decision to administer chemotherapy, targeted therapy, immunotherapy, and/or combined with surgery or radiotherapy.
Whether to treat or not to treat the disease is always a question in daily practice. Clinicians would like to know the true relationship between the observations and interventions (inputs) and the results (outputs). In other words, to find a model for disease detection, classification, or prediction. Currently, this knowledge is based on clinical trials and the experience of doctors. This exhausts the doctors in reading images and/or pathology slides repeatedly to make an accurate diagnosis. Reviewing charts to determine the best treatment options for patients also consumes a considerable amount of time. A good prediction/classification model would simplify the entire process. Here, artificial intelligence(AI) is introduced.
AI is a general term that does not have a strict definition. AI is an algorithm driven by existing data to predict or classify objects [8]. The main components include the dataset used for training, pretreatment method, an algorithm used to generate the prediction model, and the pre-trained model to accelerate the speed of building models and inherit previous experience. Machine learning (ML) is a subclass of AI, and is the science of obtaining algorithms to solve problems without being explicitly programmed, including decision trees (DTs), support vector machines (SVMs), and Bayesian networks (BNs). Deep learning is a further subclass of ML, featured with multiple layered ML, achieving feature selection and model fitting at the same time [9]. The hierarchical relationship between those definitions is displayed in Figure 1.
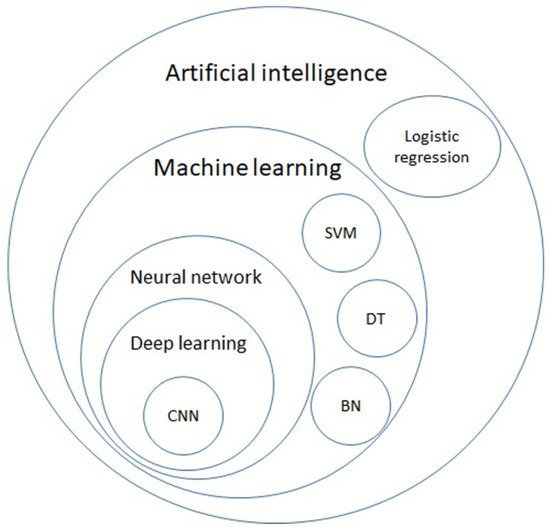
Figure 1. Venn diagram of artificial intelligence (AI), machine learning (ML), neural network, deep learning, and further algorithms in each category. AI is a general term for a program that predicts an answer to a certain problem, where one of the conventional methods is logistic regression. ML learns the algorithm through input data without explicit programming. ML includes algorithms such as decision trees (DTs), support vector machines (SVMs), and Bayesian networks (BNs). By using each ML algorithm as a neuron with multiple inputs and a single output, a neural network is a structure that mimics the human brain. Deep learning is formed with multiple layers of neural networks, and convolutional neural network (CNN) is one of the elements of the famous architecture.
2. Diagnosis
When a nodule is detected, clinicians must know the properties of the lung nodule. The gold standard is to acquire tissue samples via either biopsy or surgery. The image features provide a way to guess the properties of the lung nodule by radiomics as mentioned in the previous section. Aside from imaging features, the histopathological features also affect further treatment. Following the path of digital radiology, whole slide imaging (WSI) has opened the trend of digital histopathology. With digitalized WSI data, AI can help pathologists with daily tasks and beyond, ranging from tumor cell recognition and segmentation [10], histological subtype classification [11][12][13][14], PD-L1 scoring [15], to tumor-infiltrating lymphocyte (TIL) count [16].
2.1. Radiomics
Following the idea of radiomics in nodule detection and malignancy risk stratification, radiomics was applied to predict the histopathological features of lung nodules/masses [17]. Researchers used logistic regression of radiomics and clinical features to distinguish small cell lung cancer from non-small cell lung cancer with an AUC of 0.94 and an accuracy of 86.2% [18]. The LASSO logistic regression model was used to classify adenocarcinomas and squamous cell carcinomas in the NSCLC group [19]. Further molecular features such as Ki-67 [20], epidermal growth factor receptor (EGFR) [21], anaplastic lymphoma kinase (ALK) [22], and programmed cell death 1 ligand, (PD-L1) [23] were also shown to be predictable with AI-analyzed radiomics, a non-invasive and simple method.
2.2. WSI
The emergence of WSI is a landmark in modern digital pathology. The WSI depends on a slide scanner that can transform glass slides into digital images with the desired resolution. Once the images are stored on the server, pathologists can view them on their personal computers or handheld devices. Similar to DICOM in diagnostic radiology, in 2017, the FDA approved two vendors for the WSI system for primary diagnosis [24][25]. Meanwhile, the DICOM also planned support for WSI in the PACS systems to facilitate the adaption of digital pathology in hospitals and further information exchange [26][27]. These features enable the building of a digital pathology network to share expertise for consultations and make education across the country possible [28].
Each WSI digital slide is a large image. It may contain more than 4 billion pixels and may exceed 15 GB when scanned with a resolution of 0.25 micrometers/pixel, referred to as 40× magnification [27][29]. With recent advances in AI and DL in image classification, segmentation, and transformation, digitalized WSI provides another broad field to play. There are many applications for deep learning in cytopathology.
2.3. Histopathology
Detecting cancerous regions is the most basic and essential task of deep learning in pathology. Some models combine the detection, segmentation, and histological subtyping together [10][11][12]. Accuracy depends on the data quality, quantity, and abundance of the malignant cell differentiation status. It is difficult to perform histological subtyping of lung cancer without special immunohistochemistry (IHC) staining. This causes inter-observer disagreement when reading H&E staining. While the agreement between pathologists came to a Kappa value of 0.485, a trained AI model can achieve a Kappa value of up to 0.525 when compared with a pathologist [11]. In the detection of lymph node metastasis, a well-trained AI model can help reduce human workload and prevent errors [30]. It obviously performs better than a pathologist in a limited time and has a greater detection rate of single-cell metastasis or micro-metastasis [30].
Although WSI with H&E-stained slides is designed to view the morphology of tissues, with the aid of AI, researchers have designed methods to predict specific gene mutations, PD-L1 expression level, treatment response, and even the prognosis of patients. Focusing on lung adenocarcinoma, Coudray et al. developed an AI application using Inception-V3 for the prediction of frequently appearing gene mutations including STK11, EGFR, FAT1, SETBP1, KRAS, and TP53 [13]. The AUC of this prediction reached 0.754 for EGFR and 0.814 for KRAS which can be treated with effective targeted agents. Sha et al., used ResNet-18 as the backbone to predict the PD-L1 status in NSCLC [31]. Their model showed an AUC between 0.67 and 0.81, while different PD-L1 cutoff levels were chosen. They believed that the morphological features may be related to PD-L1 expression level.
Next-generation sequencing (NGS) plays an important role in modern lung cancer treatment [32]. Successful NGS testing depends on a sufficient number of tumor cells and tumor DNA. AI can assist in determining tumor cellularity [33][34]. In addition, a trained AI can help count the immune cells, while the tissue specimen is adequately stained for special surface markers [16]. Since the PD-L1 expression level is the key predictor for immunotherapy in lung cancer, AI has been trained to count the proportion score for PD-L1 expression [15][35]. When properly stained, computer-aided PD-L1 scoring and quantitative tumor microenvironment analysis may meet the requests of pathologists, and eliminate inter-observer variations and achieve precise lung cancer treatment [36].
However, there are several barriers to the translation of AI applications into clinical services. First, AI applications may not work well when applied to other pathology laboratories, scanners, or diverse protocols [37]. Second, most AIs are designed for their own unique functions. Users are requested to launch several applications for different purposes and spend a lot of time transferring the data. Medical devices powered by AI applications require approval by regulations. Most articles and works were in-house studies and laboratory-developed tests. All of these barriers may restrict the deployment of trained AI models in daily clinical practice [28].
2.4. Cytology
The WSI for cytology differs from pathology. Cytology slides are not even sliced flat layers. Instead, they have an entire cell on the glass and would be multiple cell layers. Cytologists tend to use the focus function and look into the cells. While digitalizing the cytology glass slide, the focus function was simulated through the Z-stack function and multiple layers of different focus [38][39]. This method yields a larger WSI file, approximately 10 times that of a typical histological case. Multiple image layers also increase complexity and pose challenges to AI applications.
Few articles have discussed cytology, especially those focusing on lung cancer. For thyroid cancer, Lin et al. proposed a DL method for thyroid fine-needle aspiration (FNA) samples and ThinPrep (TP) cytological slides for detecting papillary thyroid carcinoma [40]. The authors did not claim the ability to detect other cell types of thyroid cancer using their method. AI can be performed for various cytology samples from lung cancer patients, including pleural effusion, lymph node aspiration, tissue aspiration samples, and endobronchial ultrasound-guided fine-needle aspiration (EBUS-TBNA) of mediastinal lymph nodes.
3. Decision Making and Prognosis Prediction
Oncologists prefer to deploy this technique to its limits. There are many exciting possibilities for the use of the AI technique. By predicting treatment response, including survival and adverse events, AI was proven to have the potential to play a role in clinical decision making [41], to help surgeons choose the specific groups of patients to receive surgery, and to aid radiotherapists in planning the radiation zone.
3.1. Medication Selection
In late-stage lung cancer, the identification of driver mutations, PD-L1 expression, and tumor oncogenes affects most the treatment of choice. Using WSI and radiomics, AI could help to identify EGFR mutations [21][13], ALK [22], and PD-L1 expression [23][31][42]. EGFR mutation subtypes have also been classified using radiomic features [43].
Another research point is the use of radiomics, WSI, and clinical data to directly predict cancer treatment response or survival [44]. Dercle et al. retrospectively analyzed the data from prospective clinical trials and found that the AI model based on the random forest algorithm and CT-based radiomic features predicted the treatment sensitivity of nivolumab with an AUC of 0.77, docetaxel with an AUC of 0.67, and gefitinib with an AUC of 0.82 [45]. CT-based radiomics models have also been reported to predict the overall survival of lung cancer [46][47].
One patent application publication declared that using radiomics features of segmented cell nuclei of lung cancer can predict responses to immunotherapy with an AUC up to 0.65 in the validation dataset [48]. Although there is no specific survival prediction model for lung cancer, Ellery et al. developed a risk prediction model using the TCGA Pan-Cancer WSI database including lung cancer [49]. However, the DL algorithm did not provide acceptable prediction power for lung adenocarcinoma or lung squamous cell carcinoma.
3.2. Surgery
The gold standard for the treatment of early-stage lung cancer is surgical resection. AI was applied to pre-surgical evaluation [50][51], and prognosis prediction after surgery, and could help identify patients who are suitable to receive adjuvant chemotherapy after surgery [52].
In pre-surgical evaluation, radiologist-level AI could help predict visceral pleural invasion [51], and identify early stage lung adenocarcinomas suitable for sub-lobar resection [50]. After surgery, AI could play a role in predicting prognosis. The model based on radiomic feature nomograms could identify high-risk groups whose postsurgical tumor recurrence risk is 16-fold higher than that of low-risk group [53]. The CNN model pre-trained with the radiotherapy dataset successfully predict a 2-year overall survival after surgery [54]. The model integrating genomic and clinicopathological features was able to identify patients at risk for recurrence and who were suitable to receive adjuvant therapy [52].
3.3. Radiotherapy
SBRT is currently the standard of care to treat early-stage lung cancer and/or provide local control for patients who are medically inoperable or refuse surgery. Radiomics-based models have been reported to successfully predict 1-year tumor recurrence via CT scans performed after 3 and 6 months of SBRT [55]. Lewis and Kemp also developed a model trained on TCGA dataset to predict cancer resistance to radiation [56]. As a well-known side effect of radiotherapy, radiation pneumonitis can be lethal, and clinicians would like to prevent this situation. The AI model based on pretreatment CT radiomics was superior to the traditional model using dosimetric and clinical predictors in predicting radiation pneumonitis [57]. Another ANN algorithm trained with radiomics extracted from a 3D dose map of radiotherapy has been shown to predict the acute and late pulmonary toxicities with an accuracy of 0.69 [58]. A well-designed prediction model for radiation pneumonitis may help to prevent radiation pneumonitis in the future.
This entry is adapted from the peer-reviewed paper 10.3390/cancers14061370
References
- Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 29 November 2021).
- Luo, Y.H.; Chiu, C.H.; Scott Kuo, C.H.; Chou, T.Y.; Yeh, Y.C.; Hsu, H.S.; Yen, S.H.; Wu, Y.H.; Yang, J.C.; Liao, B.C.; et al. Lung Cancer in Republic of China. J. Thorac. Oncol. 2021, 16, 519–527.
- Cause of Death Statistics. Available online: https://www.mohw.gov.tw/lp-4650-2.html (accessed on 1 October 2021).
- Panunzio, A.; Sartori, P. Lung Cancer and Radiological Imaging. Curr. Radiopharm. 2020, 13, 238–242.
- Migliore, M.; Palmucci, S.; Nardini, M.; Basile, A. Imaging patterns of early stage lung cancer for the thoracic surgeon. J. Thorac. Dis. 2020, 12, 3349–3356.
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260.
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of advances since 2015. J. Thorac. Oncol. 2021, 17, 362–387.
- Klang, E. Deep learning and medical imaging. J. Thorac. Dis. 2018, 10, 1325–1328.
- Lawson, C.E.; Marti, J.M.; Radivojevic, T.; Jonnalagadda, S.V.R.; Gentz, R.; Hillson, N.J.; Peisert, S.; Kim, J.; Simmons, B.A.; Petzold, C.J.; et al. Machine learning for metabolic engineering: A review. Metab. Eng. 2021, 63, 34–60.
- Šarić, M.; Russo, M.; Stella, M.; Sikora, M. CNN-based method for lung cancer detection in whole slide histopathology images. In Proceedings of the 2019 4th International Conference on Smart and Sustainable Technologies (SpliTech), Split, Croatia, 18–21 June 2019; pp. 1–4.
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019, 9, 3358.
- Gertych, A.; Swiderska-Chadaj, Z.; Ma, Z.; Ing, N.; Markiewicz, T.; Cierniak, S.; Salemi, H.; Guzman, S.; Walts, A.E.; Knudsen, B.S. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 2019, 9, 1483.
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567.
- Wang, X.; Chen, H.; Gan, C.; Lin, H.; Dou, Q.; Tsougenis, E.; Huang, Q.; Cai, M.; Heng, P.-A. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 2019, 50, 3950–3962.
- Kapil, A.; Meier, A.; Zuraw, A.; Steele, K.E.; Rebelatto, M.C.; Schmidt, G.; Brieu, N. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci. Rep. 2018, 8, 17343.
- Aprupe, L.; Litjens, G.; Brinker, T.J.; van der Laak, J.; Grabe, N. Robust and accurate quantification of biomarkers of immune cells in lung cancer micro-environment using deep convolutional neural networks. PeerJ 2019, 7, e6335.
- Wu, G.; Jochems, A.; Ibrahim, A.; Yan, C.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Structural and functional radiomics for lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3961–3974.
- Liu, S.; Liu, S.; Zhang, C.; Yu, H.; Liu, X.; Hu, Y.; Xu, W.; Tang, X.; Fu, Q. Exploratory study of a CT Radiomics model for the classification of small cell lung cancer and non-small-cell lung cancer. Front. Oncol. 2020, 10, 1268.
- Zhu, X.; Dong, D.; Chen, Z.; Fang, M.; Zhang, L.; Song, J.; Yu, D.; Zang, Y.; Liu, Z.; Shi, J. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur. Radiol. 2018, 28, 2772–2778.
- Gu, Q.; Feng, Z.; Liang, Q.; Li, M.; Deng, J.; Ma, M.; Wang, W.; Liu, J.; Liu, P.; Rong, P. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 2019, 118, 32–37.
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986.
- Song, L.; Zhu, Z.; Mao, L.; Li, X.; Han, W.; Du, H.; Wu, H.; Song, W.; Jin, Z. Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front. Oncol. 2020, 10, 369.
- Zhu, Y.; Liu, Y.-L.; Feng, Y.; Yang, X.-Y.; Zhang, J.; Chang, D.-D.; Wu, X.; Tian, X.; Tang, K.-J.; Xie, C.-M. A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas. Ann. Transl. Med. 2020, 8, 930.
- Evans, A.J.; Bauer, T.W.; Bui, M.M.; Cornish, T.C.; Duncan, H.; Glassy, E.F.; Hipp, J.; McGee, R.S.; Murphy, D.; Myers, C. US Food and Drug Administration approval of whole slide imaging for primary diagnosis: A key milestone is reached and new questions are raised. Arch. Pathol. Lab. Med. 2018, 142, 1383–1387.
- Abels, E.; Pantanowitz, L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J. Pathol. Inform. 2017, 8, 23.
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261.
- DICOM Whole Slide Imaging (WSI). Available online: https://dicom.nema.org/Dicom/DICOMWSI/ (accessed on 29 November 2021).
- Sakamoto, T.; Furukawa, T.; Lami, K.; Pham, H.H.N.; Uegami, W.; Kuroda, K.; Kawai, M.; Sakanashi, H.; Cooper, L.A.D.; Bychkov, A. A narrative review of digital pathology and artificial intelligence: Focusing on lung cancer. Transl. Lung Cancer Res. 2020, 9, 2255.
- Giovagnoli, M.R.; Giansanti, D. Artificial Intelligence in Digital Pathology: What Is the Future? Part 1: From the Digital Slide Onwards. Healthc. Multidiscip. Digit. Publ. Inst. 2021, 9, 858.
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 2017, 318, 2199–2210.
- Sha, L.; Osinski, B.L.; Ho, I.Y.; Tan, T.L.; Willis, C.; Weiss, H.; Beaubier, N.; Mahon, B.M.; Taxter, T.J.; Yip, S.S. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 2019, 10, 24.
- Biermann, J.; Adkins, D.; Agulnik, M.; Benjamin, R.; Brigman, B.; Butrynski, J.; Cheong, D.; Chow, W.; Curry, W.; Frassica, D. National comprehensive cancer network. Bone cancer. J. Natl. Compr. Cancer Netw. 2013, 11, 688–723.
- Furukawa, T.; Kuroda, K.; Bychkov, A.; Pham, H.; Kashima, Y.; Fukuoka, J. Verification of Deep Learning Model to Measure Tumor Cellularity in Transbronchial Biopsies of Lung Adenocarcinoma; Laboratory Investigation, Nature Publishing Group: New York, NY, USA, 2019.
- Sakamoto, T.; Furukawa, T.; Pham, H.H.; Kuroda, K.; Tabata, K.; Kashima, Y.; Okoshi, E.N.; Morimoto, S.; Bychkov, A.; Fukuoka, J. Collaborative workflow between pathologists and deep learning for evaluation of tumor cellularity in lung adenocarcinoma. bioRxiv 2022.
- Hondelink, L.M.; Hüyük, M.; Postmus, P.E.; Smit, V.T.; Blom, S.; von der Thüsen, J.H.; Cohen, D. Development and validation of a supervised deep learning algorithm for automated whole-slide programmed death-ligand 1 tumour proportion score assessment in non-small cell lung cancer. Histopathology 2021, 80, 635–647.
- Wu, J.; Lin, D. A Review of Artificial Intelligence in Precise Assessment of Programmed Cell Death-ligand 1 and Tumor-infiltrating Lymphocytes in Non− Small Cell Lung Cancer. Adv. Anat. Pathol. 2021, 28, 439–445.
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309.
- Giansanti, D.; Grigioni, M.; D’Avenio, G.; Morelli, S.; Maccioni, G.; Bondi, A.; Giovagnoli, M.R. Virtual microscopy and digital cytology: State of the art. Ann. Dell’istituto Super. Di Sanità 2010, 46, 115–122.
- Boschetto, A.; Pochini, M.; Bottini, L.; Giovagnoli, M.R.; Giansanti, D. The focus emulation and image enhancement in digital cytology: An experience using the software Mathematica. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2015, 3, 110–116.
- Lin, Y.-J.; Chao, T.-K.; Khalil, M.-A.; Lee, Y.-C.; Hong, D.-Z.; Wu, J.-J.; Wang, C.-W. Deep Learning Fast Screening Approach on Cytological Whole Slides for Thyroid Cancer Diagnosis. Cancers 2021, 13, 3891.
- Sesen, M.B.; Nicholson, A.E.; Banares-Alcantara, R.; Kadir, T.; Brady, M. Bayesian networks for clinical decision support in lung cancer care. PLoS ONE 2013, 8, e82349.
- Jiang, M.; Sun, D.; Guo, Y.; Guo, Y.; Xiao, J.; Wang, L.; Yao, X. Assessing PD-L1 expression level by radiomic features from PET/CT in nonsmall cell lung cancer patients: An initial result. Acad. Radiol. 2020, 27, 171–179.
- Li, S.; Ding, C.; Zhang, H.; Song, J.; Wu, L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019, 46, 4545–4552.
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696.
- Dercle, L.; Fronheiser, M.; Lu, L.; Du, S.; Hayes, W.; Leung, D.K.; Roy, A.; Wilkerson, J.; Guo, P.; Fojo, A.T. Identification of non–small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin. Cancer Res. 2020, 26, 2151–2162.
- Le, V.-H.; Kha, Q.-H.; Hung, T.N.K.; Le, N.Q.K. Risk score generated from CT-based radiomics signatures for overall survival prediction in non-small cell lung cancer. Cancers 2021, 13, 3616.
- Sun, F.; Chen, Y.; Chen, X.; Sun, X.; Xing, L. CT-based radiomics for predicting brain metastases as the first failure in patients with curatively resected locally advanced non-small cell lung cancer. Eur. J. Radiol. 2021, 134, 109411.
- Predicting Response to Immunotherapy Using Computer Extracted Featuresof Cancer Nuclei from Hematoxylin and Eosin (H&E) Stained Images of Non-Small Cell Lung Cancer (NSCLC). Available online: https://patft.uspto.gov/netacgi/nph-Parser?Sect1=PTO2&Sect2=HITOFF&p=1&u=%2Fnetahtml%2FPTO%2Fsearchbool.html&r=1&f=G&l=50&co1=AND&d=PTXT&s1=11,055,844.PN.&OS=PN/11,055,844&RS=PN/11,055,844 (accessed on 6 March 2022).
- Wulczyn, E.; Steiner, D.F.; Xu, Z.; Sadhwani, A.; Wang, H.; Flament-Auvigne, I.; Mermel, C.H.; Chen, P.-H.C.; Liu, Y.; Stumpe, M.C. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE 2020, 15, e0233678.
- Yoshiyasu, N.; Kojima, F.; Hayashi, K.; Bando, T. Radiomics technology for identifying early-stage lung adenocarcinomas suitable for sublobar resection. J. Thorac. Cardiovasc. Surg. 2021, 162, 477–485.e1.
- Choi, H.; Kim, H.; Hong, W.; Park, J.; Hwang, E.J.; Park, C.M.; Kim, Y.T.; Goo, J.M. Prediction of visceral pleural invasion in lung cancer on CT: Deep learning model achieves a radiologist-level performance with adaptive sensitivity and specificity to clinical needs. Eur. Radiol. 2021, 31, 2866–2876.
- Jones, G.D.; Brandt, W.S.; Shen, R.; Sanchez-Vega, F.; Tan, K.S.; Martin, A.; Zhou, J.; Berger, M.; Solit, D.B.; Schultz, N. A genomic-pathologic annotated risk model to predict recurrence in early-stage lung adenocarcinoma. JAMA Surg. 2021, 156, e205601.
- D’Antonoli, T.A.; Farchione, A.; Lenkowicz, J.; Chiappetta, M.; Cicchetti, G.; Martino, A.; Ottavianelli, A.; Manfredi, R.; Margaritora, S.; Bonomo, L. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 2020, 27, 497–507.
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711.
- Mattonen, S.A.; Palma, D.A.; Haasbeek, C.J.; Senan, S.; Ward, A.D. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Med. Phys. 2014, 41, 033502.
- Lewis, J.E.; Kemp, M.L. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat. Commun. 2021, 12, 2700.
- Krafft, S.P.; Rao, A.; Stingo, F.; Briere, T.M.; Court, L.E.; Liao, Z.; Martel, M.K. The utility of quantitative CT radiomics features for improved prediction of radiation pneumonitis. Med. Phys. 2018, 45, 5317–5324.
- Bourbonne, V.; Da-Ano, R.; Jaouen, V.; Lucia, F.; Dissaux, G.; Bert, J.; Pradier, O.; Visvikis, D.; Hatt, M.; Schick, U. Radiomics analysis of 3D dose distributions to predict toxicity of radiotherapy for lung cancer. Radiother. Oncol. 2021, 155, 144–150.
This entry is offline, you can click here to edit this entry!
