Enterococci are part of the subdominant microbiota of many artisanal cheeses. As a genus that has evolved for resistance against adverse environmental factors and that readily exchanges genetic elements, enterococci are well adapted to the cheese environment and may reach high numbers in artisanal cheeses. Their metabolites impact cheese flavor, texture, and rheological properties, thus contributing to the development of its typical sensorial properties. Due to their antimicrobial activity, enterococci modulate the cheese microbiota, stimulate autolysis of other lactic acid bacteria (LAB), control pathogens and deterioration microorganisms, and may offer beneficial effects to the health of their hosts. They could in principle be employed as adjunct/protective/probiotic cultures; however, due to their propensity to acquire genetic determinants of virulence and antibiotic resistance, together with the opportunistic character of some of its members, this genus does not possess Qualified Presumption of Safety (QPS) status. It is, however, noteworthy that some putative virulence factors described in foodborne enterococci may simply reflect adaptation to the food environment and to the human host as commensal. Further research is needed to help distinguish friend from foe among enterococci, eventually enabling exploitation of the beneficial aspects of specific cheese-associated strains.
- dairy Enterococcus
- starter culture
- adjunct culture
- protective culture
- probiotics
- opportunistic pathogen
- pathogenicity determinants
- antimicrobial resistance
1. Introduction
Enterococci are part of the subdominant microbiota of many artisanal cheeses [1], where they play an important role in the development of sensorial properties [1], help modulating the cheese microbiota [2] and may play a role in controlling pathogenic as well as deterioration microorganisms [3]. Their evolutionary history has primed them to resist (harsh conditions), adapt (to stress), and persist (in difficult environments). It has endowed this genus of commensal gut bacteria with plastic genomes and a notable capacity to trade genes by horizontal transfer events, both of a homologous and heterologous nature [4]. The adaptation to their hosts provided them the opportunity to become important opportunistic pathogens [4]; the ability to easily acquire genes by horizontal transfer allowed them to build up a considerable resistome [5], as well as several putative determinants of virulence [6]. On the other hand, enterococcal strains have been used as adjunct cultures, probiotics and may have beneficial effects upon several aspects of their human hosts’ health [7]. The impact of the enterococcal microbiota of artisanal cheeses on the health of their hosts is still the object of debate, despite the wealth of knowledge gathered in recent years on their presence, technological properties, potential health benefits, antibiotic resistance, and carriage of virulence factors [7]. Due to the uncertainty on their safety, enterococci do not possess QPS status in the EU and are not Generally Regarded as Safe (GRAS) in the USA. The lack of a recognized safety status as a genus has hampered their use as industrial food cultures, despite their potential benefits [8]. A better perception of their roles in the cheese system is required to both eventually allow for their industrial use and to understand their potential role as reservoirs of genetic determinants of antibiotic resistance and virulence.
2. The Genus Enterococcus—A Bacterial Group That Has Evolved for Resistance
As a genus, enterococci have a relatively short history, most of which intertwined with that of other Gram-positive cocci, especially streptococci. The terms “streptococci” and “enterococci” first appeared during the Golden Age of Microbiology. “Streptococcos” has been employed for the first time in 1874, by Billroth [9], a contemporary of Louis Pasteur, to describe cocci arranged in chains that he observed in wounds. A decade later, streptococci were described as a genus, in Rosenbach’s work on bacteria from suppurated wounds [10]. The first mention to the term “enterococcus” occurred less than 20 years later, when Thiercellin & Jouhaud [11] reported isolating saprophytic, potentially pathogenic cocci from the human intestines. These new cocci were then thought to be part of the Streptococcus genus, and received the species epithet of faecalis [12]. Thirty years later, when Rebecca Lancefield published her serological classification of streptococci [13] and, subsequently, when James Sherman proposed division of streptococci into four groups [14], enterococci emerged as a separate group of “streptococcal lineages”; however, they would not be afforded the status of genus on their own until the mid-80s, when, based on molecular biology approaches, Karl Schleifer and Renate Kilpper-Bälz [15] proposed that the Gram-positive cocci belonging to Lancefield’s group D constitute, in fact, a genus, and named it after Thiercellin & Jouhaud’s cocci. At that time, only two species—Enterococcus faecalis and Enterococcus faecium—were assigned to the newly proposed genus. Presently, at the doorstep of the third decade of the new millennium, it encompasses close to 60 validly published species with correct name [16].
The association between enterococci and the human intestinal habitat has been established since the early works of Thiercellin & Jouhaud [11]. More recently, research by Lebreton et al. [4][17] has shown that enterococci are indeed a native enteric genus that has colonized from there a variety of human- and non-human-related habitats. Their ability to colonize these very diversified ecological niches stemmed from their evolutionary history, which has been hypothesized to have started 500 ± 130.5 Myr, driven by terrestrialization of the animal hosts that sheltered the enterococcal ancestors [17]. Based on 16S rRNA data, enterococci seem to have arisen from Vagococcus-like ancestors, which in turn stemmed out of the Carnobacteriacea branch of the bacterial tree of life. Carnobacteria and Vagococcus remained associated with marine hosts [17], whereas most enterococci relate to the gut of terrestrial animals (mammals, birds, reptiles, and insects) [17][18][19]. As a result of their evolution towards hardiness, members of this genus have been found in soils [20][21], waters [22], plants [21][23], hospital environments [24], foods [8][25] and feeds [26][27], further to their association with human and animal microbiota. Enterococci do not sporulate; hence, their persistence in these seemingly hostile environments stems, in part, from their ability to enter a viable, non-culturable state [28], their notable resistance to starvation, and their capacity to withstand desiccation [17]. The plasticity of the enterococcal genome granted them not only the ability to colonize a wide range of niches, but also capacitated them to play a wide range of roles in these environments—as well as in their human hosts—where they range from harmless commensals, through opportunistic pathogens, to probiotic bacteria [29].
3. Raw Milk as a Source of Enterococci for Dairy Products
Lactic acid bacteria are part of the core, mesophilic/psychrotrophic microbiota of raw bovine milk [30][31]. Although lactococci, lactobacilli, and streptococci may reach populations in the order of 104 CFU mL−1 in raw milk, the numbers of leuconostocs and enterococci reach only 103 CFU mL−1 [30].
Enterococci may be present to large numbers in dairy products (up to 108 CFU g−1) [32]. They are among the most common lactic acid bacteria in raw milk [24], which they access from dairy environment, animals, and humans [32][33][34][35]. Despite their relation to the intestinal microbiota of humans and dairy animals, fecal contamination does not seem to play an important role upon entrance of enterococci into the dairy production chain. The milking equipment has instead been regarded as their major source in raw milk [36]. The work by Gelsomino et al. [34] has demonstrated that the milking machine and the bulk tank are important sources of enterococci for milk and dairy products. The milking environment was found to be the source of vancomycin-resistant enterococci (VRE), rather than the animals [37]. Mastitis can also be a source of multidrug-resistant enterococci [38]. The season of the year may also play a role in modulating the enterococcal diversity in raw milk [39].
Raw milk may, therefore, serve as a source of enterococci for dairy products, even when pasteurization is applied during processing a posteriori. There are indeed reports of enterococci surviving pasteurization temperatures [40]. Their thermal resistance, however, is highly variable and species-dependent, with z values ranging from 5.0 (E. faecalis) to 9.8 °C (E. hirae) [41]. The thermal resistance of enterococci depends also on the growth phase and previous thermal history of the cells [42]; hence, the enterococcal microbiota in pasteurized milk differs from that of raw milk. Although Enterococcus durans tends to predominate in the former, the most prevalent enterococcal species in the latter is E. faecalis [39]. Besides their thermoduric character, post-treatment recontamination also explains the presence of enterococci in pasteurized milk products, such as cheese [32], with biofilms on milk-contact surfaces acting as a potential source of high numbers of bacteria [43][44].
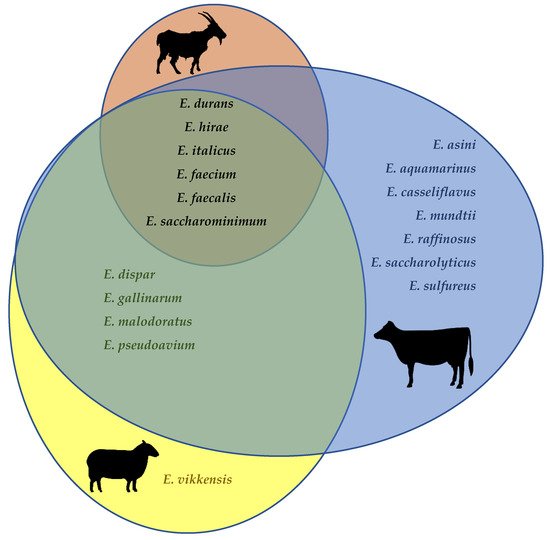
Enterococcus faecalis, E. faecium, E. durans, E. hirae, as well as (albeit infrequently) E. saccharominimum and E. italicus, have been isolated from the raw milk of cows, goats, and sheep (Figure 1) [31][36][38][39][41][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64]. Additionally, E. dispar, E. malodoratus, E. pseudoavium and E. gallinarum have been isolated both in cow’s and in ewe’s milk. E. casseliflavus, E. mundtii, E. aquamarinus, E. asini, E. saccharolyticus, E. sulfureus, and E. raffinosus have been described only in cow’s milk, whereas E. vikkensis was found just in ewe’s milk. It is worth keeping in mind, however, that the apparently lesser diversity of enterococcal species in goat’s and sheep’s milk when compared to cow’s milk may merely reflect the much fewer studies that focused on the former.
Several of the enterococcal species described in raw milk (e.g., E. faecalis, E. faecium and E. mundtii) are known to include strains that produce bacteriocins. Therefore, enterococci might play a role in modulating the milk microbiota, as well as the bacterial communities within dairy products, thus contributing both to their safety and to their sensorial properties [65]. Several enterococcal species from raw milk have demonstrated important technological properties with impact upon the sensorial properties of dairy products, such as diacetyl production, autolytic activity, proteolytic activity [31][66] and lipolytic activity [67], as well as probiotic potential [68][69].
This entry is adapted from the peer-reviewed paper 10.3390/foods10040821
References
- Casey, M.G.; Häni, J.-P.; Gruskovnjak, J.; Schaeren, W.; Wechsler, D. Characterisation of the non-starter lactic acid bacteria (NSLAB) of Gruyère PDO cheese. Lait 2006, 86, 407–414.
- Fox, P.F.; Cogan, T.M.; Guinee, T.P. Chapter 52. Factors That Affect the Quality of Cheese. In Cheese: Chemistry, Physics and Microbiology, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 617–641.
- Franz, C.M.A.P.; Van Belkum, M.J.; Holzapfel, W.H.; Abriouel, H.; Gálvez, A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 2007, 31, 293–310.
- Lebreton, F.; Willems, R.J.L.; Gilmore, M. Enterococcus diversity, origins in nature, and gut colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 5–63.
- Werner, G.; Coque, T.M.; Franz, C.M.; Grohmann, E.; Hegstad, K.; Jensen, L.; van Schaik, W.; Weaver, K. Antibiotic resistant enterococci–tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 2013, 303, 360–379.
- Arias, C.; Murray, B. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278.
- Graham, K.; Stack, H.; Rea, R. Safety, beneficial and technological properties of enterococci for use in functional food applications–a review. Crit. Rev. Food Sci. Nutr. 2020, 60, 3836–3861.
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: Between probiotic potential and safety concerns–an update. Front. Microbiol. 2018, 9, 1791.
- Billroth, A.W. Untersuchungen über die Vegetationsformen von Coccobacteria Septica; Georg Reimer: Berlin, Germany, 1874.
- Rosenbach, F.J. Mikro-Organismen bei den Wund-Infections-Krankheiten des Menschen; Bergmann, J.F., Ed.; Verlag from J.F. Bergmann: Wiesbaden, Germany, 1884.
- Thiercelin, E.; Jouhaud, L. Reproduction de l’entérocoque; taches centrales; granulations peripheriques et microblastes. Comptes Rendus Seances Soc. Biol. Paris 1903, 55, 686–688.
- Andrews, F.W.; Horder, T.J. A study of streptococci pathogenic for man. Lancet 1906, 2, 708–713.
- Lancefield, R.C. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 1933, 57, 571–595.
- Sherman, J.M. The enterococci and related streptococci. J. Bacteriol 1937, 35, 81–93.
- Schleifer, K.H.; Kilpper-Bälz, R. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol. 1984, 34, 31–34.
- DSMZ. LPSN-List of Prokaryotic Names with Standing in Nomenclature. 2020. Available online: https://lpsn.dsmz.de/ (accessed on 10 February 2021).
- Lebreton, F.; Manson, A.L.; Saavedra, J.T.; Straub, T.J.; Earl, A.M.; Gilmore, M.S. Tracing the enterococci from Paleozoic origins to the hospital. Cell 2017, 169, 849–861.
- Mundt, J.O. Occurrence of enterococci in animals in a wild environment. J. Appl. Microbiol. 1963, 11, 136–140.
- Martin, J.D.; Mundt, J.O. Enterococci in insects. J. Appl. Microbiol. 1972, 24, 575–580.
- Silva, V.; Peixoto, F.; Igrejas, G.; Parelho, C.; Garcia, P.; Carvalho, I.; Sousa, M.; Pereira, J.E.; Rodrigues, A.; Poeta, P. First report on vanA-Enterococcus faecalis recovered from soils subjected to long-term livestock agricultural practices in Azores Archipelago. Int. J. Environ. Res. 2018, 12, 39–44.
- Ben Said, L.B.; Klibi, N.; Dziri, R.; Borgo, F.; Boubadous, A.; Ben Slama, K.; Torres, C. Prevalence, antimicrobial resistance and genetic lineages of Enterococcus spp. from vegetable food, soil and irrigation water in farm environments in Tunisia. J. Sci. Food Agric. 2016, 96, 1627–1633.
- Moore, D.F.; Guzman, J.A.; McGee, C. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. J. Appl. Microbiol. 2008, 105, 1017–1025.
- Müller, T.; Ulrich, A.; Ott, E.-M.; Müller, M. Identification of plant-associated enterococci. J. Appl. Microbiol. 2001, 91, 268–278.
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82.
- Ben Braiek, O.; Smaoui, S. Enterococci: Between Emerging Pathogens and Potential Probiotics. Biomed. Res. Int. 2019, 2019, 5938210.
- Novais, C.; Campos, J.; Freitas, A.R.; Barros, M.; Silveira, E.; Coque, T.M.; Antunes, P.; Peixe, L. Water supply and feed as sources of antimicrobial-resistant Enterococcus spp. in aquacultures of rainbow trout (Oncorhyncus mykiss), Portugal. Sci. Total Environ. 2018, 625, 1102–1112.
- Channaiah, L.H.; Subramanyam, B.; Zurek, L. Molecular characterization of antibiotic resistant and potentially virulent enterococci isolated from swine farms and feed mills. J. Stored Prod. Res. 2018, 77, 189–196.
- Lleò, M.M.; Bonato, B.; Benedetti, D.; Canepari, P. Survival of enterococcal species in aquatic environments. FEMS Microbiol. Ecol. 2005, 54, 189–196.
- Zhong, Z.; Kwok, L.-Y.; Hou, Q.; Sun, Y.; Li, W.; Zhang, L.; Sun, Z. Comparative genomic analysis revealed great plasticity and environmental adaptation of the genomes of Enterococcus faecium. BMC Genom. 2019, 20, 60.
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698.
- Perin, L.M.; Miranda, R.O.; Todorov, S.D.; Franco, B.D.G.; Nero, L.A. Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int. J. Food Microbiol. 2014, 185, 121–126.
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2003, 26, 163–171.
- Garg, S.K.; Mital, B.K. Enterococci in milk and milk products. Crit. Rev. Microbiol. 1991, 18, 15–45.
- Gelsomino, R.; Vancanneyt, M.; Cogan, T.M.; Condon, S.; Swings, J. Source of enterococci in a farmhouse raw-milk cheese. Appl. Environ. Microbiol. 2002, 68, 3560–3565.
- Kakgli, D.M.; Vancanneyt, M.; Vandamme, P.; Hill, C.; Cogan, T.M. Contamination of milk by enterococci and coliforms from bovine faeces. J. Appl. Microbiol. 2007, 103, 1393–1405.
- Kakgli, D.M.; Vancanneyt, M.; Hill, C.; Vandamme, P.; Cogan, T.M. Enterococcus and Lactobacillus contamination of raw milk in a farm dairy environment. Int. J. Food Microbiol. 2007, 114, 243–251.
- Ortigosa, M.; Irigoyen, A.; Urdin, M.; García, S.; Ibañez, F.C.; Torre, P. Sources of enterococci in Idiazábal-type cheese. Int. J. Food Microbiol. 2008, 125, 146–152.
- Wu, X.; Hou, S.; Zhang, Q.; Ma, Y.; Zhang, Y.; Kan, W.; Zhao, X. Prevalence of virulence and resistance to antibiotics in pathogenic enterococci isolated from mastitic cows. J. Vet. Med. Sci. 2016, 78, 1663–1668.
- McAuley, C.M.; Britz, M.L.; Gobius, K.S.; Craven, H. Prevalence, seasonality, and growth of enterococci in raw and pasteurized milk in Victoria, Australia. J. Dairy Sci. 2015, 98, 8348–8358.
- Giraffa, G.; Carminati, D.; Neviani, E. Enterococci isolated from dairy products: A review of risks and potential technological use. J. Food Prot. 1997, 60, 732–738.
- McAuley, C.; Gobius, K.S.; Britz, M.L.; Craven, H.M. Heat resistance of thermoduric enterococci isolated from milk. Int. J. Food Microbiol. 2012, 154, 162–168.
- Martínez, S.; López, M.; Bernardo, A. Thermal inactivation of Enterococcus faecium: Effect of growth temperature and physiological state of microbial cells. Lett. Appl. Microbiol. 2003, 37, 475–481.
- Didienne, R.; Defargues, C.; Callon, C.; Meylheuc, T.; Hulin, S.; Montel, M.-C. Characteristics of microbial biofilm on wooden vats (‘gerles’) in PDO Salers cheese. Int. J. Food Microbiol. 2012, 156, 91–101.
- Jamet, E.; Akary, E.; Poisson, M.-A.; Chamba, J.-F.; Bertrand, X.; Serror, P. Prevalence and characterization of antibiotic resistant Enterococcus faecalis in French cheeses. Food Microbiol. 2012, 31, 191–198.
- Bouymajane, A.; Filali, F.R.; Oulghazi, S.; Ed-dra, A.; Benhallam, F.; El Allaoui, A.; Anissi, K.; Ouhmidou, B.; Moumni, M. Occurrence, molecular and antimicrobial resistance of Enterococcus spp. isolated from raw cow’s milk trade by street trading in Meknes city, Morocco. Germs 2018, 9, 77–84.
- Garroni, E.; Doulgeraki, A.I.; Pavli, F.; Spiteri, D.; Valdramidis, V.P. Characterization of indigenous lactic acid bacteria in cow milk of the Maltese Islands: A geographical and seasonal assessment. Microorganisms 2020, 8, 812.
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154.
- Giannino, M.L.; Marzotto, M.; Dellaglio, F.; Feligni, M. Study of microbial diversity in raw milk and fresh curd used for Fontina cheese production by culture-independent methods. Int. J. Food Microbiol. 2009, 130, 188–195.
- Masoud, W.; Vogensen, F.K.; Lillevang, S.; Al-Soud, W.A.; Sørensen, S.J.; Jakobsen, M. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int. J. Food Microbiol. 2012, 153, 192–202.
- Von Neubeck, M.; Baur, C.; Krewinkel, M.; Stoeckel, M.; Kranz, B.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015, 211, 57–65.
- Alves, P.I.; Martins, M.P.; Semedo, T.; Marques, J.J.F.; Tenreiro, R.; Crespo, M.T.B. Comparison of phenotypic and genotypic taxonomic methods for the identification of dairy enterococci. Antonie van Leeuwenhoek 2003, 85, 237–252.
- Citak, S.; Yucel, N.; Mendi, A. Antibiotic resistance of enterococcal isolates in raw milk. J. Food Process. Preserv. 2015, 29, 183–195.
- Fricker, M.; Skånseng, B.; Rudi, K.; Stessl, B.; Ehling-Schulz, M. Shift from farm to dairy tank milk microbiota revealed by a polyphasic approach is independent from geographical origin. Int. J. Food Microbiol. 2011, 145, S24–S30.
- Jiménez, E.; Ladero, V.; Chico, I.; Maldonado-Barragán, A.; López, M.; Martín, V.; Fernández, L.; Fernández, M.; Álvarez, M.A.; Torres, C.; et al. Antibiotic resistance, virulence determinants and production of biogenic amines among enterococci from ovine, feline, canine, porcine and human milk. BMC Microbiol. 2013, 13, 288.
- Garnica, M.L.; Sáez-Nieto, J.A.; González, R.; Santos, J.A.; Gonzalo, C. Diversity of gram-positive catalase-negative cocci in sheep bulk tank milk by comparative 16S rDNA sequence analysis. Int. Dairy J. 2014, 34, 142–145.
- Ariznabarreta, A.; Gonzalo, C.; San Primitivo, F. Microbiological quality and somatic cell count of ewe milk with special reference to staphylococci. J. Dairy Sci. 2002, 85, 1370–1375.
- Ruiz, P.; Pérez-Martín, F.; Seseña, S.; Palop, M.L. Seasonal diversity and safety evaluation of enterococci population from goat milk in a farm. Dairy Sci. Technol. 2016, 96, 359–375.
- Cocolin, L.; Foschino, R.; Comi, G.; Fortina, M.G. Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol. 2007, 24, 752–758.
- Perin, L.M.; Todorov, S.D.; Nero, L.A. Investigation of genes involved in nisin production in Enterococcus spp. strains isolated from raw goat milk. Antonie van Leeuwenhoek 2016, 109, 1271–1280.
- Perin, L.M.; Nero, L.A. Antagonistic lactic acid bacteria isolated from goat milk and identification of a novel nisin variant Lactococcus lactis. BMC Microbiol. 2014, 14, 36.
- Júnior, W.L.G.A.; Ferrari, I.S.; Souza, J.V.; Silva, C.D.A.; Costa, M.M.; Dias, F.S. Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 2015, 53, 96–103.
- Sarkar, S.L.; Hossain, M.I.; Monika, S.A.; Sanyal, S.K.; Roy, P.C.; Hossain, M.A.; Jahid, I.K. Probiotic potential of Pediococcus acidilactici and Enterococcus faecium isolated from indigenous yogurt and raw goat milk. Microbiol. Biotechnol. Lett. 2020, 48, 276–286.
- Achemchem, F.; Cebrián, R.; Abrini, J.; Martínez-Bueno, M.; Valdivia, E.; Maqueda, M. Antimicrobial characterization and safety aspects of the bacteriocinogenic Enterococcus hirae F420 isolated from Moroccan raw goat milk. Can. J. Microbiol. 2012, 58, 596–604.
- Callon, C.; Duthoit, F.; Delbès, C.; Ferrand, M.; Le Frileux, Y.; De Crémoux, R.; Montel, M.-C. Stability of microbial communities in goat milk during a lactation year: Molecular approaches. Syst. Appl. Microbiol. 2007, 30, 547–560.
- Perin, L.M.; Pereira, J.G.; Bersot, L.S.; Nero, L.A. The microbiology of raw milk. In Raw Milk: Balance between Hazards and Benefits; Nero, L.A., Carvalho, A.F., Eds.; Academic Press: Chennai, India, 2019; pp. 45–64.
- Franciosi, E.; Settanni, L.; Cavazza, A.; Poznanski, E. Biodiversity and technological potential of wild lactic acid bacteria from raw cow’s milk. Int. Dairy J. 2009, 19, 3–11.
- Hantsis-Zacharov, E.; Halpern, M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168.
- Banwo, K.; Sanni, A.; Tan, H. Technological properties and probiotic potential of Enterococcus faecium strains isolated from cow milk. J. Appl. Microbiol. 2012, 114, 229–241.
- Elkenany, R.M.; Elsayed, M.M.; Eltaysh, R.A.; Zakaria, A.I.; El-Baz, A.H. In vitro probiotic characteristics of Enterococcus species isolated from raw cow milk. Int. J. Probiotics Prebiotics 2018, 13, 117.
