Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Developmental Biology
The CB is a membrane-less perinuclear organelle present in male germ cells which serve as storehouse for mRNAs transported by RNA binding and transport proteins like GRTH/DDX25. It also serves as a processing center of mRNAs awaiting translation during later stages of spermatogenesis. These CBs are involved in diverse pathways like RNA transport, decay, surveillance and regulate the stability of mRNAs to secure the correct timing of protein expression at different stages of spermiogenesis.
- phospho-GRTH
- chromatoid bodies
- spermatogenesis
- small RNAs
- siRNAs
- miRNAs
- PiRNAs
1. Introduction
Spermatogenesis is a complex, serial and highly specialized differentiation program during which progenitor cells undergo a series of cellular reorganization events, resulting in the production of mature functional sperm [1][2][3]. This differentiation process is controlled by the integrated expression of an array of genes and specific proteins in a precise temporal sequence that produces genetically unique spermatozoa [2][3][4]. Gene expression in haploid spermatids requires temporal uncoupling of transcription and translation in the adult mammalian testis [2][5][6]. Post-meiotic haploid round spermatids formed during the spermatogenesis process possess complex transcriptomes, and hence efficient and accurate quality control mechanisms are necessary to deal with the major diversity of transcribed RNAs in these germ cells [7]. Initial stages of spermatogenesis are marked by active transcription and the resulting mRNAs are transported and stored transiently in large cytoplasmic ribonucleoprotein granules called “chromatoid bodies” (CBs) [8][9].
The CB is a perinuclear organelle and a ribonucleoprotein (RNP) granule present in the cytoplasm of male germ cells [9]. The functions of CBs overlap with those of P-bodies and stress granules of somatic cells. Due to the presence of a wide array of proteins involved in different steps of RNA metabolism with different classes of RNAs, including microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs), the CB seems to function as an RNA processing center [9][10]. CBs are involved in diverse pathways and mechanisms through which it regulates a wide variety of processes, including RNA transport, regulation, decay, surveillance, translational arrest, translation machinery assembly, etc. Although interesting aspects have been found in the last half-decade, several intrinsic mechanisms (specifically, the role of CBs in the storage and regulation of important mRNAs that are involved in making healthy sperm) are still not fully understood in detail [10]. Mature mRNAs are transported to CBs during the early stages of spermatogenesis (active transcription stages) bound to RNA binding/transport proteins such as gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) [9][11][12].
2. RNPs Are Critical for Cellular and Organism Function: Role of CBs
RNP complexes and granules are powerful composite structures of merged functions and unique properties. RNP granules are formed under physiological conditions. In addition, granule formation can be induced by stress such as heat, starvation, oxidative stress, etc. These granules are more dynamic in nature, and they occur in condensed or diffused states and depend on the cell requirements. Specific mutations occurring in the cell can prevent RNP assembly or disassembly and diminish cellular and organism function, resulting in several pathologies. Examples include fragile X-syndrome, associated primary ovarian insufficiency in human germ cells, and amyotrophic lateral sclerosis (a progressive neurodegenerative disease that affects nerve cells in the brain and the spinal cord), possibly occurring due to the failure of disassembly of stress granules. The importance of RNPs is diverse and the complete functions are still not completely understood. The main functions of RNPs include carrying out intricate tasks in RNA-processing pathways and the post-transcriptional regulation of mRNAs. The association of RNA-binding proteins such as DDX4 and RNA in the germ line of several organisms gives rise to non-membranous structures called germ granules. These are known by several different names in different organisms with minor variations in function, from Nuage, Pole plasm, piNG body (piRNA nuage giant body) in Drosophila, Polar granule in C elegans, and chromatoid body, Balbiani body, and Cajal body in mice and zebrafish [13]. One of the biggest RNP granules is the CB, which consists primarily of RNA and other RNPs involved in the RNA processing of male germ cells.
The CB is a membrane-less, filamentous-lobular, perinuclear organelle (0.5–1 μm) and an electron-dense structure present in the cytoplasm of male germ cells [9][14]. It acts as a repository of important mRNAs associated as mRNPs waiting for translation during later stages of spermatogenesis. CBs dynamically move within the cytoplasm and exhibit continuous changes in shape and size. The biochemical composition of CBs is unique: they are primarily composed of small RNAs, Piwi-interacting RNA (piRNA), siRNAs, miRNAs, mRNAs, long non-coding RNAs, RNA-binding proteins, members of the small interfering RNA pathway such as MIWI, Argonaute protein, Dicer endonuclease, decapping enzyme, and other proteins involved in RNA post-transcriptional regulation which may play a critical role during sperm elongation and spermiogenesis [8][9][15][16]. CBs also contain GRTH [9], phospho-GRTH [10] and MVH/DDX4, a mouse homolog of VASA, a germ cell marker [16]. In fact, these proteins constitute the majority of CBs in addition to other proteins which are involved in the piRNA pathway, nonsense-mediated RNA decay (NMD) pathway, and in RNA post-transcriptional and translational regulation. Specifically, these CB proteins are DDX25, DDX4, MILI, MIWI, TDRD6, TDRD7, D1PAS1, PABP1, HSPA2, and constitute around 70% of the CB proteome [9][10][17][18].
The origin of CBs is highly debatable. The most accepted view is that they emerged from small granules (precursors of CBs) which are associated with the nuclear envelope present near the electron-dense inter-mitochondrial cement in the late pachytene spermatocytes (Figure 1). The CB can be detected during all steps of round spermatid differentiation (steps 1–8 of spermiogenesis). The largest CBs are observed in step four, five and six of spermiogenesis [18]. At later stages of spermatid elongation, CBs move caudally to the neck region and split into two separate structures; one is discarded along with residual cytoplasm, and the other forms a ring around the base of the flagellum (Figure 1).
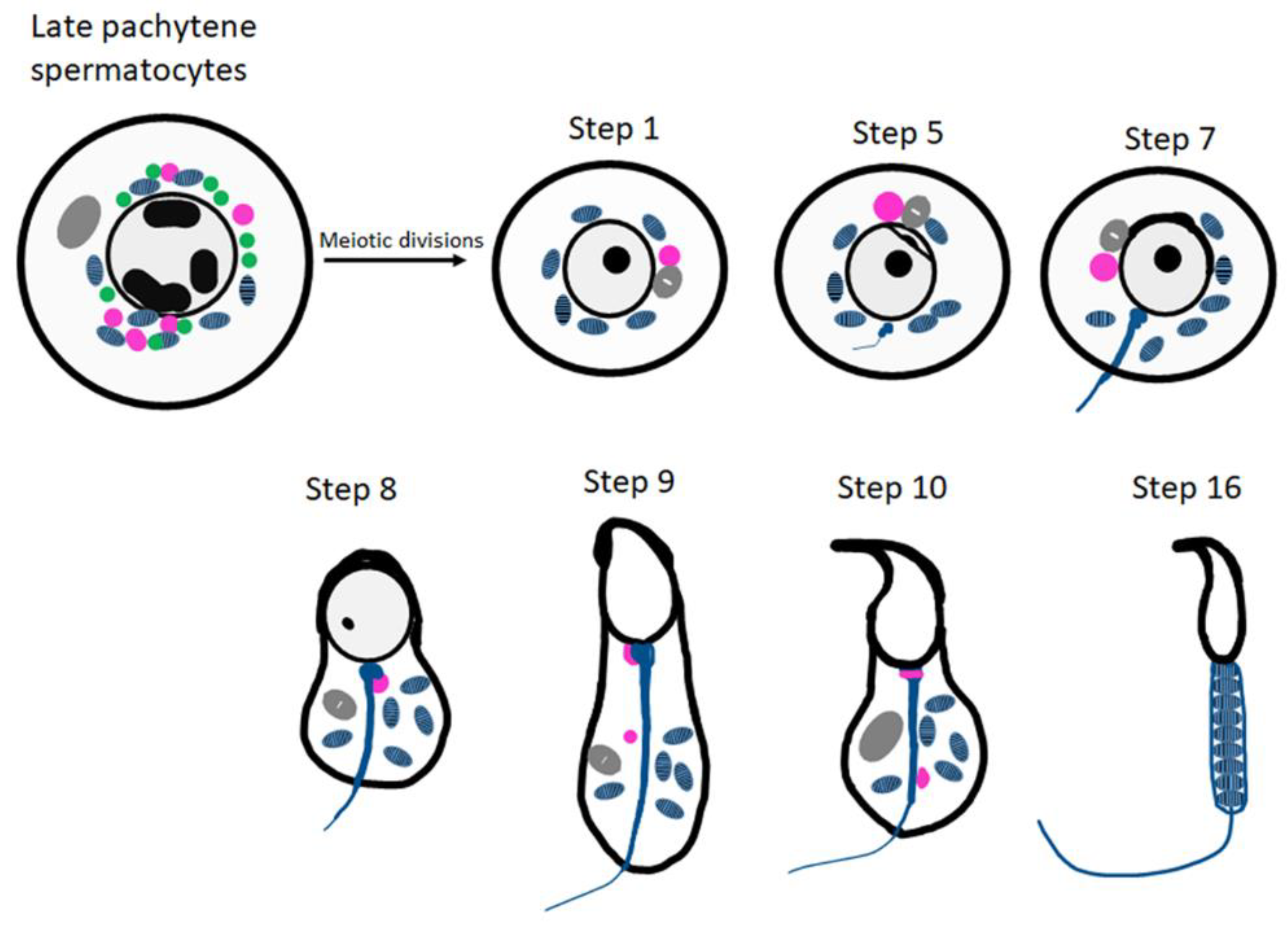
Figure 1. Schematic representation of CB organization and fate during spermiogenesis in mice. The inter-mitochondrial cement (IMC; green) intermixed with mitochondria (blue) and the CB precursors (pink) co-exist in late pachytene spermatocytes. The Golgi complex is depicted in gray. The CB (pink) is condensed to its final single form in the early round spermatids. At step 8 of spermiogenesis, the CB is found at the basis of the flagellum. Later, it splits into two separate structures (step 9 onwards) and eventually disappears.
There is limited information about the specific mechanisms of CB function during spermatid elongation. With the identification of small non-coding RNA-mediated gene regulation and other associated mechanisms, the functions of the CB are slowly beginning to be understood. However, several of the molecular mechanisms are still unclear, requiring further in-depth molecular studies.
3. CBs Are Analogous to Stress Granules and P-Bodies
Compartmentalization of molecular processes is accomplished by various intracellular organelles that spatially segregate functionally related molecules. RNPs act as organelles that lack any demarcating membrane and play a key role in mRNA homeostasis. RNP granules formed under physiological conditions in male germ cells are called CBs, while in somatic cells they are called RNA processing bodies or P-bodies. CBs’ overall functions fall between P-bodies and stress granules and are in concert with the maintenance of RNA regulation. Stress granules and processing bodies are also membrane-less RNA granules that dynamically sequester translationally inactive messenger ribonucleoprotein particles (mRNPs) into compartments which are distinct from the surrounding cytoplasm [19][20]. These granules are more dynamic in nature and exist in a condensed or diffused state based on conditions and requirements. Like P-bodies, stress granule assembly is dependent on the pool of non-translating mRNAs. Stress granules and P-bodies can physically interact to facilitate the shuttling of RNA and protein between them. The main difference between P-bodies and stress granules is that P-bodies assemble around the key enzymes of cytoplasmic RNA degradation in physiological conditions, and stress granules assemble around the essential components of the translation machinery under different stress conditions such as heat, glucose deprivation, viral or bacterial infection, hypoxia, and oxidative stress [19][21].
In contrast to CBs and stress granules, P-bodies are not associated with the regulation of translation initiation; instead they serve as a site for mRNA degradation, translation repression, storage of non-translating mRNAs, and RNA-binding proteins (Figure 2) [10][20][22]. P-bodies are uniquely enriched with factors related to mRNA decay and the NMD pathway, such as members of mRNA decapping machinery including the decapping enzymes DCP1/2; UPF1/2, the activators of decapping EDC3, Dhh1/RCK/p54, Pat1, Scd6/RAP55, and EDC3; the LSM1-7 complex; and the exonuclease XRN1 (Figure 2) [20][22][23][24]. P-bodies are independent of initiation factors or translational assembly, while CBs seem to regulate mRNA storage and decay, as well as translational machinery, all in one compartment effectively [10][20]. Recent studies have shown that the process of liquid–liquid phase separation is a main driver promoting the assembly of RNP granules such as stress granules, P-bodies, and other related membrane-less organelles [25]. It would be very interesting to determine the involvement of biological mediators and other proteins in the assembly of RNP granules. Few studies have implicated post-translational modifications of RNP granule proteins as the driver for the assembly and disassembly of the RNP granule [13][19]. Irrespective of the exact driver which initiates the formation of the RNPs, the role it plays is unique and irreplaceable, and has a critical role in the survival of the cell. More like stress granules, CBs have proteins or mRNA that are involved in the translation process, such as initiation and elongation factors, ribosomal subunits and other associated factors, and mRNA regulatory factors such as PABPC1, UPF2, and others [10][17][21]. RNP granules induced by heat stress are detected in spermatogonia, preleptotene, and early pachytene spermatocytes [26]. Detailed information and similarities of these heat-induced RNP granules in germ cells with respect to CBs are not clear. Further studies on interaction between these RNPs inside the cell provide vital clues on the molecular mechanisms of gene regulation. CBs resemble the recently described TIS-associated granules and the interconnections proposed between TIS granules and the ER (TIGER domain structure) [27].
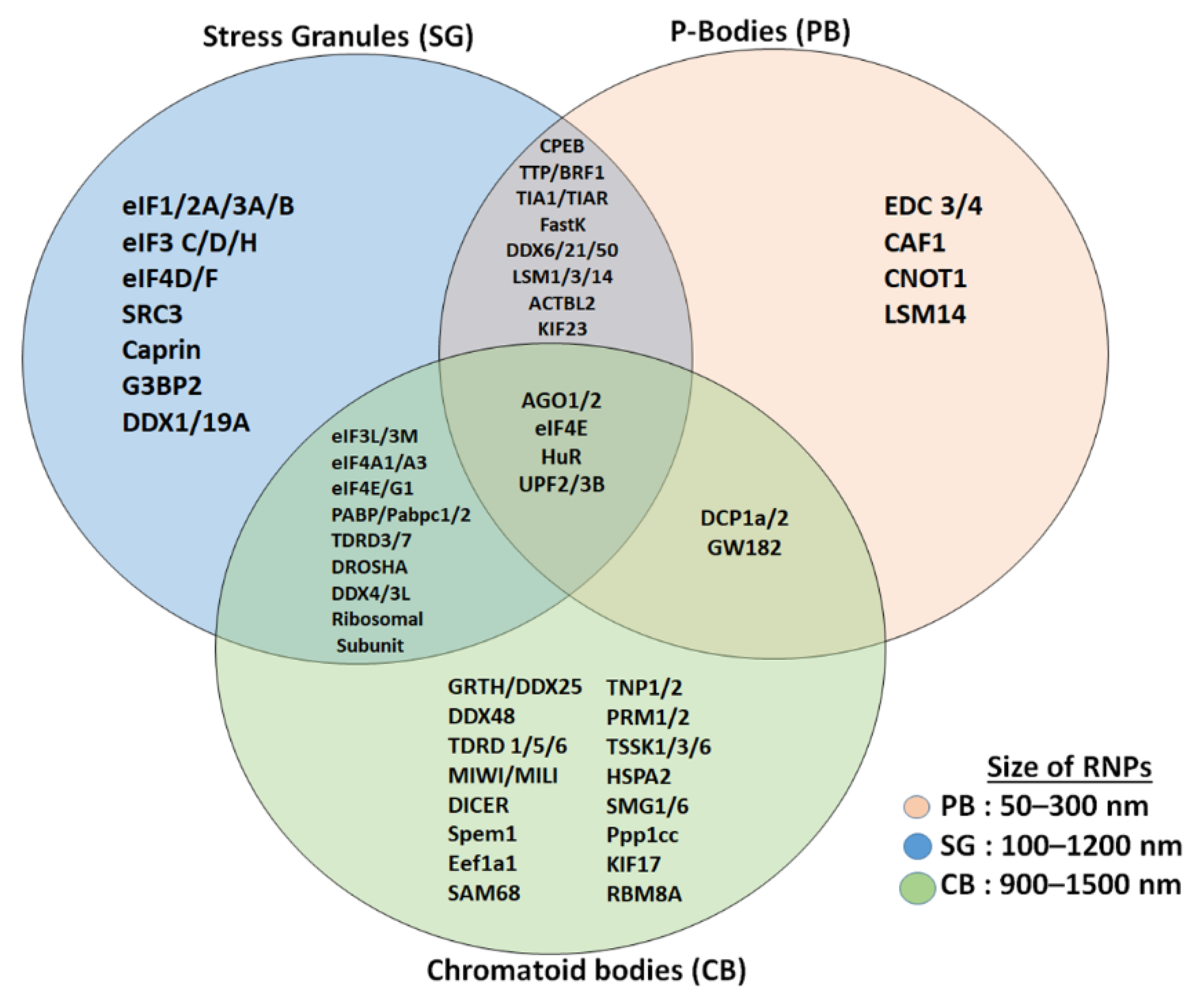
Figure 2. Comparison of proteins (mRNA decay and translation machinery) present in CBs with stress granules and P-bodies. Stress granules and CBs share several initiation factors and translation initiation assembly proteins, while P-bodies and CBs share few of the proteins. CBs are more related to stress granules than to P-bodies and CBs are bigger in size compared to stress granules and P-bodies.
4. Small RNAs of CBs and Regulation of Spermatogenesis
During spermatogenesis, the early precursors of sperm utilize small regulatory RNAs such as microRNAs to control the expression of an array of genes at transcriptional or post-transcriptional levels during complex and specialized processes of sperm production. The role of CBs in small RNA-mediated gene control and the associated mechanisms are not clearly delineated at present. The repertoires of microRNAs and piRNAs have been identified and both serve as important regulators of male germ cell differentiation.
miRNAs are small in size (21–23 nucleotides) and belong to the class of noncoding RNAs which act as endogenous gene regulators and participate in a wide array of biological functions, by promoting target mRNA degradation and inhibition of translation [28][29][30]. Each miRNA can target several mRNAs of different genes and thus regulates gene expression stringently in a stage- and tissue-specific manner in every organ including the testis. miRNAs recognize their target mRNAs by sequence-specific bp pairing in the RNA-induced silencing complex together with Argonaute proteins (AGO) [9]. Most miRNAs are derived from primary miRNA transcripts which are processed by the Drosha-DGCR8 complex in the nucleus to generate precursor miRNAs (pre-miRNAs), which are transported to the cytoplasm where mature miRNAs are generated via a Dicer-dependent or independent route. One of the crucial components of the miRNA and siRNA pathways is the cytoplasmic endonuclease Dicer, which is critical for male fertility [31]. Dicer interacts with the germ-cell-specific RNA helicase MVH (mouse VASA homolog). Sertoli cell-specific deletion of Dicer in mice results in spermatogenic malfunction, defective maturation, and infertility [31]. Transcripts of AGO proteins, Drosha, and Dicer have been demonstrated to be present in germ cells and Sertoli cells [29][32]. GRTH regulates proteins of the microprocessor complex, Drosha and DGCR8 (miRNA biogenesis) at the mRNA and protein levels [29]. miRNA pathway proteins have been demonstrated to accumulate in the CBs in haploid round spermatids, suggesting that the CB and GRTH have a role in miRNA-dependent gene regulation [28][29].
Testis-specific miRNAs such as miR-469, testis-preferred miR-34c, miR-470 and others such as let-7 family members (let-7a/d, b, and e-g), and miR203 were upregulated in round spermatids of GRTH KO mice. Furthermore, the enzyme complex (Drosha-DGCR8) required for the processing of miRNA was also upregulated in the KO mice. MiR-469 target TP2 and Prm2 mRNA by binding to their coding regions and thereby preventing translation of these essential mRNAs to proteins [29]. GRTH negatively regulates overall miRNA biogenesis via Drosha/DGCR8 microprocessor complex, to generate mature miR-469 and others which could play a role during spermatogenesis. miRNAs such as miR-469, through their inhibitory action on TP2/Prm2 mRNA, control the timely expression for chromatin compaction and the progression of spermatogenesis [29].
piRNAs comprise the biggest and most complex class of small non-coding RNAs. Unlike miRNAs (22 nt long), piRNAs are slightly longer, in the range of 26–32 nucleotides, and the biosynthesis of piRNAs is different from that of miRNAs and siRNAs [33][34]. During post-meiotic germ cell differentiation, the CB accumulates piRNAs and proteins of piRNA machinery, as well as several other proteins involved in distinct RNA regulation pathways. The embryonic and post-natal male germ cells express high levels of piRNAs during late meiotic cells and haploid round spermatids. CBs provide platforms for the Piwi-interacting RNA (piRNA) pathway and appear to be involved both in piRNA biogenesis and piRNA-targeted RNA degradation. In addition, RNA regulatory mechanisms, such as the NMD pathway, are also known to exist inside the CB, and this provides exciting new insights into the function of CBs. Another important and most fascinating feature of the CB is its dynamic and non-random movements in the cytoplasm of haploid spermatids, thereby facilitating the sharing of selective mRNAs, small RNAs and proteins between neighboring spermatids, and making them an attractive organelle for specific pathways’ interconnectivity, in addition to coordinating mRNA regulation in an efficient and accurate manner.
This entry is adapted from the peer-reviewed paper 10.3390/cells11040613
References
- Steger, K. Haploid spermatids exhibit translationally repressed mRNAs. Anat. Embryol. 2001, 203, 323–334.
- Kimmins, S.; Sassone-Corsi, P. Chromatin remodelling and epigenetic features of germ cells. Nature 2005, 434, 583–589.
- Griswold, M.D. Spermatogenesis: The Commitment to Meiosis. Physiol. Rev. 2016, 96, 1–17.
- Kavarthapu, R.; Anbazhagan, R.; Raju, M.; Morris, C.T.; Pickel, J.; Dufau, M.L. Targeted knock-in mice with a human mutation in GRTH/DDX25 reveals the essential role of phosphorylated GRTH in spermatid development during spermatogenesis. Hum. Mol. Genet. 2019, 28, 2561–2572.
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; Griswold, M.D. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004, 71, 319–330.
- Tsai-Morris, C.H.; Sheng, Y.; Gutti, R.; Tang, P.Z.; Dufau, M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25): A multifunctional protein essential for spermatogenesis. J. Androl. 2010, 31, 45–52.
- Maclean, J.A.; Wilkinson, M.F. Gene regulation in spermatogenesis. Curr. Top. Dev. Biol. 2005, 71, 131–197.
- Kotaja, N.; Sassone-Corsi, P. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007, 8, 85–90.
- Sato, H.; Tsai-Morris, C.H.; Dufau, M.L. Relevance of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the structural integrity of the chromatoid body during spermatogenesis. Biochim. Biophys. Acta 2010, 1803, 534–543.
- Anbazhagan, R.; Kavarthapu, R.; Dufau, M.L. Role of phosphorylated gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the regulation of germ cell specific mRNAs in chromatoid bodies during spermatogenesis. Front. Cell Dev. Biol. 2020, 8, 580019.
- Sheng, Y.; Tsai-Morris, C.H.; Gutti, R.; Maeda, Y.; Dufau, M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J. Biol. Chem. 2006, 281, 35048–35056.
- Dufau, M.L.; Tsai-Morris, C.H. Gonadotropin-regulated testicular helicase (GRTH/DDX25): An essential regulator of spermatogenesis. Trends Endocrinol. Metab. 2007, 18, 314–320.
- Schisa, J.A.; Elaswad, M.T. An emerging role for post-translational modifications in regulating rnp condensates in the germ line. Front. Mol. Biosci. 2021, 8, 658020.
- Peruquetti, R.L. Perspectives on mammalian chromatoid body research. Anim. Reprod. Sci. 2015, 159, 8–16.
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22.
- Meikar, O.; Da Ros, M.; Korhonen, H.; Kotaja, N. Chromatoid body and small RNAs in male germ cells. Reproduction 2011, 142, 195–209.
- Meikar, O.; Vagin, V.V.; Chalmel, F.; Sõstar, K.; Lardenois, A.; Hammell, M.; Jin, Y.; Da Ros, M.; Wasik, K.A.; Toppari, J.; et al. An atlas of chromatoid body components. RNA 2014, 20, 1–3.
- Lehtiniemi, T.; Kotaja, N. Germ granule-mediated RNA regulation in male germ cells. Reproduction 2018, 155, 77–91.
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J. Cell. Biol. 1999, 147, 1431–1442.
- Riggs, C.L.; Kedersha, N.; Ivanov, P.; Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 2020, 133, jcs242487.
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 16, 871–884.
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032813.
- Parker, R.; Sheth, U. P Bodies and the Control of mRNA Translation and Degradation. Mol. Cell 2007, 25, 635–646.
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286.
- Guo, L.; Shorter, J. It’s raining liquids: RNA tunes viscoelasticity and dynamics of membraneless organelles. Mol. Cell 2015, 60, 189–192.
- Kim, B.; Cooke, H.J.; Rhee, K. DAZL is essential for stress granule formation implicated in germ cell survival upon heat stress. Development 2012, 139, 568–578.
- Ma, W.; Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3’utr-mediated protein-protein interactions. Cell 2018, 175, 1492–1506.
- Kotaja, N.; Bhattacharyya, S.N.; Jaskiewicz, L.; Kimmins, S.; Parvinen, M.; Filipowicz, W.; Sassone-Corsi, P. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. USA 2006, 103, 2647–2652.
- Dai, L.; Tsai-Morris, C.H.; Sato, H.; Villar, J.; Kang, J.H.; Zhang, J.; Dufau, M.L. Testis-specific miRNA-469 up-regulated in gonadotropin-regulated testicular RNA helicase (GRTH/DDX25)-null mice silences transition protein 2 and protamine 2 messages at sites within coding region: Implications of its role in germ cell development. J. Biol. Chem. 2011, 286, 44306–44318.
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610.
- Papaioannou, M.D.; Pitetti, J.L.; Ro, S.; Park, C.; Aubry, F.; Schaad, O.; Vejnar, C.E.; Kühne, F.; Descombes, P.; Zdobnov, E.M.; et al. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev Biol. 2009, 326, 250–259.
- Gonzalez-Gonzalez, E.; Lopez-Casas, P.P.; del Mazo, J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim. Biophys. Acta 2008, 1779, 306–311.
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007, 316, 744–747.
- Thomson, T.; Lin, H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu. Rev. Cell Dev. Biol. 2009, 25, 355–376.
This entry is offline, you can click here to edit this entry!
