Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
In plants, salicylic acid (SA) is a hormone that mediates a plant’s defense against pathogens. SA also takes an active role in a plant’s response to various abiotic stresses, including chilling, drought, salinity, and heavy metals. In addition, in recent years, numerous studies have confirmed the important role of SA in plant morphogenesis.
- salicylic acid
- root growth
- lateral roots
- adventitious roots
- abiotic stress
- plant defense
- auxin
1. Introduction
Recognized as the sixth plant hormone in 1992 [1], 2-hydroxybenzoic or salicylic acid (SA) belongs to a family of naturally occurring phenolic compounds which possess an aromatic benzene ring bearing one or more hydroxyl groups. Since then, a huge amount of data has been accumulated on SA’s involvement in various biological processes. As reviewed in [2][3], SA has secured a reputation as a vital defense hormone. At the same time, SA’s impact on cell, tissue, and organ phenotypes is well established (reviewed in [4]). Despite the growing evidence that SA is an important growth regulator, its morphogenetic role, especially in relation to roots, has rarely been summarized in reviews.
It is worthy of note that, in roots, SA content and its dynamic during development may differ from that in shoots [5][6][7][8], which can potentially cause differences in SA functions. For instance, SA basal level in shoots is 2–100 times higher than in roots, depending on the species [5][6]. The ratio between free and conjugated SA forms, also differs [6]. For example, the shoots of wheat seedlings, three days after germination (DAG), contain about 48 times more free than conjugated SA, whereas in the roots, the contrary is seen, with conjugated SA levels exceeding the level of free SA by about six times. During wheat seedling growth, SA content in both free and conjugated forms gradually decreases in shoots but not in roots. In 14 DAG seedlings, the conjugated SA becomes prevalent in both organs but the ratio of free to conjugated form still differs slightly and amounts to 0.4 and 0.5 for shoots and roots, respectively. These differences provide ample reason to consider the role of SA in root morphology, distinct from its function in shoots.
The phenotypic analysis of SA deficient/accumulating lines and SA-treated plants provides insight into the role of SA in plant growth and development. However, the data on changes in root morphology in SA mutants are scarce and often contradictory. For example, decreased root length is reported in SA-accumulating Arabidopsis mutants [9][10][11] and in SA-depleted rice mutants [12][13]. In rice, the inhibitory effects of an SA deficiency on root length have been reported in relation to both SA biosynthesis mutant aim1 [12] and plants transgenic for the bacterial Naphthalene hydroxylase G (NahG) gene, encoding salicylate hydroxylase that inactivates SA by converting it to catechol [13]. In contrast, transgenic Lotus japonicus plants expressing NahG, demonstrate enhanced root growth [14]. These contradictions may be due to species-specific basal SA levels, which vary greatly between plant species, even those belonging to the same family [4][15].
2. SA Metabolism and Signaling in Plants
SA metabolism has been comprehensively described in numerous reviews, for example, [16][17][18][19][20]; therefore, the researchers touch only briefly on this aspect in this review. SA is synthesized in plants, bacteria, and fungi from chorismate, the final product in the shikimate pathway (reviewed in [16]). Chorismate is also the primary source for the biosynthesis of aromatic amino acids (tryptophan, phenylalanine, and tyrosine) and a wide range of aromatic secondary metabolites, including flavonoids, alkaloids, and lignins. SA biosynthesis (Figure 1) starts in plastids, where chorismate is converted into either isochorismate via isochorismate synthase (ICS) or prephenate via chorismate mutase (CM), giving rise to two parallel ICS and phenylalanine ammonia-lyase (PAL) pathways of SA biosynthesis (reviewed in [16][17][19][20]). The relative contributions of the ICS and PAL pathways to SA biosynthesis are species-dependent with an equal contribution being made in soybean and a prevalence of ICS and PAL pathways being seen in Arabidopsis and rice, respectively. In the ICS pathway, ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) transports isochorismate to the cytosol, where it is conjugated with glutamate by avrPphB Susceptible 3 (PBS3) to produce isochorismate-9-glutamate, which is either spontaneously decomposed into SA or converted to SA by an acyltransferase Enhanced Pseudomonas Susceptibility 1 (EPS1). In the PAL pathway, there are two ways of prephenate elaboration into phenylalanine (reviewed in [18]). In plastids, prephenate aminotransferases (PPA-ATs) catalyze its transition to arogenate, which is then converted by arogenate dehydratase (ADT) into phenylalanine. In the cytosol, the prephenate–phenylalanine transition is realized through phenylpyruvate by prephenate dehydratase (PDT) and phenylpyruvate aminotransferase (PPY-AT). PAL turns phenylalanine into trans-cinnamic acid, following which the formation of SA occurs either through ortho-coumaric acid or benzoic acid (reviewed in [16][17][19][20]). In the latter case, Abnormal Inflorescence Meristem 1 (AIM1), a 3-hydroxyacyl-CoA dehydrogenase, contributes to this process. Benzoic acid is hydroxylated to SA, possibly by benzoic acid 2-hydroxylase (BA2H).
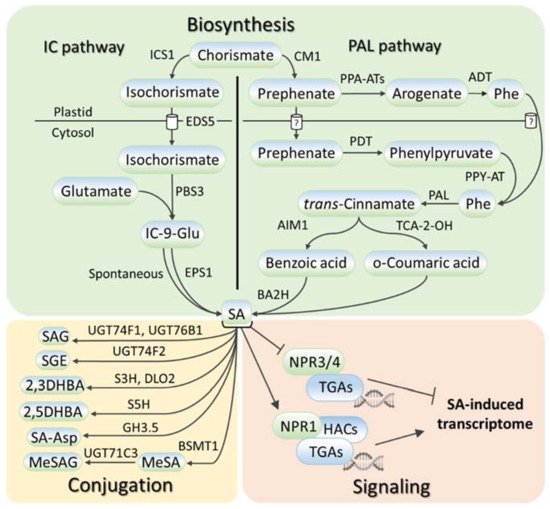
Figure 1. SA metabolism and signaling in plants. SA is synthesized via two routes, the isochorismate pathway or the phenylalanine ammonia-lyase pathway, which both start with chorismate. SA conversions include SA glycosylation, methylation, hydroxylation, and amino-acid conjugation. SA signaling depends on the interaction of SA receptor NPR1 with TGA transcription factors and histone acetyltransferases. SA, salicylic acid; ICS1, isochorismate synthase 1; EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5; PBS3, avrPphB Susceptible 3; EPS1, Enhanced Pseudomonas Susceptibility 1; IC-9-Glu, isochorismate-9-glutamate; CM1, chorismate mutase 1; PPA-ATs, prephenate aminotransferases; PDT, prephenate dehydratase; PPY-AT, phenylpyruvate aminotransferase; ADT, arogenate dehydratase; PAL, phenylalanine ammonia-lyase; AIM1, Abnormal Inflorescence Meristem 1; TCA-2-OH, trans-cinnamic acid 2-hydroxylase; BA2H, benzoic acid 2-hydroxylase; UGT74F1/74F2/76B1/71C3, UDP-glucosyltransferases 74F1, 74F2, 76B1 and 71C3; S5H, SA-5 hydroxylase; S3H, SA-3 hydroxylase; DLO2, DMR6-LIKE OXYGENASE 2; GH3.5, Gretchen Hagen 3.5; BSMT1, benzoic acid/salicylic acid methyltransferase; SAG, salicylic acid 2-O-β-D-glucose; SGE, salicylic acid glucose ester; 2,3/2,5-DHBA, 2,3/2,5-dihydroxybenzoic acid; SA-Asp, salicyloyl-L-aspartate; MeSA, methyl salicylate; MeSAG, methyl salicylate O-β-glucoside; NPR1/3/4, NONEXPRESSOR OF PATHOGENESIS RELATED GENES 1/3/4; HACs, histone acetyltransferases; TGA, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN.
A wide number of regulators control SA biosynthesis (Table S1). Among them, the reactive oxygen species (ROS), particularly hydrogen peroxide, form a self-amplifying feedback loop with SA, in which hydrogen peroxide promotes SA biosynthesis, and SA induces hydrogen peroxide accumulation by inactivating its scavengers [21][22] (reviewed in [23]).
SA levels are regulated not only by SA biosynthesis but also by SA chemical modifications and intercellular transport (reviewed in [18][24]). These processes have been studied mainly in Arabidopsis. SA glycosylation occurs via the conjugation of glycosyl onto the hydroxyl and carboxyl groups of SA, producing two inactive SA storage forms, salicylic acid 2-O-β-d-glucose (SAG) and salicylic acid glucose ester (SGE). Uridine diphosphate (UDP)-glycosyltransferases UGT74F1 and UGT74F2 perform the conversion to the former, while only UGT74F2 is involved in the catalysis to the latter. The carboxyl group can be also methylated by the S-adenosyl-L-methionine (SAM)-dependent methyltransferase, BA/SA carboxyl methyltransferase 1 (BSMT1), producing methyl salicylate (MeSA), the form of SA that has increased membrane permeability. SA hydroxylation by SA-5 and SA-3 hydroxylases generates 2,3-DHBA and 2,5-DHBA dihydroxybenzoic acids. Gretchen Hagen 3.5/WESO 1 (GH3.5/WES1) and another unknown GH3 family enzyme convert SA into salicyloyl-L-aspartate (SA-Asp). Some of these conjugated forms of SA may also be glycosylated. Inactive SA forms can be stored until they are required to increase the active pool of free SA; alternatively, some of them may be subjected to SA catabolism. SA is often spread via apoplast (reviewed in [24]). Since SA is a weak acid with poor water solubility, the existence of influx and efflux carriers along with pH-dependent diffusion is proposed for its movement through the plasma membrane.
The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES (NPR) are the SA receptors (reviewed in [3][18][24]). At a low SA level, NPR1 oligomerizes in the cytosol. Meanwhile, NPR1 paralogs, NPR3 and NPR4, directly interact with the basic leucine zipper (bZIP) family’s TGA transcription factors on the promoters of NPR1 targets, to suppress their expression. SA facilitates the reduction of cytosolic NPR1 oligomers into monomers, which are translocated to the nucleus and activate transcription in complex with TGAs. At the same time, SA inhibits the activity of NPR3 and NPR4 to allow for the transcription of SA-responsive genes. The NPR1 pathway is functional in both shoots and roots [25][26]. SA also binds to A subunits of protein phosphatase 2A (PP2A) and inhibits the activity of this enzyme, thereby altering auxin transport and distribution [26]. There are other SA binding proteins as well but their functions in SA signaling are largely unknown [27][28][29][30] (reviewed in [31][32]).
3. Modulation of Endogenous SA Levels in Roots
In Arabidopsis shoots, the basal level of SA amounts to 0.25–1 µg per gram of the fresh weight, rising up to 20 µg.g−1 at the place of pathogen attack [33] (reviewed in [4]). In many plant species roots also accumulate SA upon invasion of soil-borne pathogens (Table 1). Rapid SA accumulation is a part of plant immune signaling, which has been extensively studied in shoots. In this process, SA promotes pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), effector-triggered immunity (ETI), and systemic acquired resistance (SAR), via an NPR-dependent activation of plant defense genes, to resist biotrophic and semi-biotrophic pathogens (reviewed in [2][3][34]). The mechanisms of plant defense in roots are less studied, yet pathogen-induced SA accumulation is considered an essential factor in root protection from biotic stress [35] (reviewed in [36]). The attacks of soil-borne pathogens are capable of inducing systemic SA accumulation in above-ground tissue [37], and mutants and transgenic plants with a reduced ability to accumulate SA are more susceptible to root infections than wild types [38][39][40][41].
Table 1. The influence of biotic and abiotic stress factors on SA content in roots.
| Plant Species | Stress Factor Type | Stress Factor 1 | SA Level | Reference |
|---|---|---|---|---|
| Biotic stress | ||||
| Cucumus sativus L. | Necrotrophic fungus | Rhizoctonia solani | ↑ | [37] |
| Zea mays L. | Root herbivore | Diabrotica virgifera larvae | ↑ | [42] |
| Arabidopsis thaliana L. (Bur-0) | Biotrophic protist | Plasmodiophora brassicae | ↑ | [43] |
| Arabidopsis thaliana L. (Col-0) | Biotrophic protist | Plasmodiophora brassicae | - | [43] |
| Abiotic stress | ||||
| Cassia tora L. | Aluminium | Al (10–50 µM) | ↑ (RT) | [44] |
| Glycine max L. | Aluminium | AlCl3 (30 μM) | ↑ (RT) | [45] |
| Hordeum vulgare L. | Heavy metal | CdCl2 (25 µM) | ↑ (F) | [46] |
| Triticum aestivum L. | Heavy metal | Cd(NO3)2 (250 µM) | ↑ (F) | [47] |
| Arabidopsis thaliana L. (Col) | Heavy metal | CdCl2 (50 μM) | ↑ | [48] |
| Oryza sativa L. | Chilling | 5 °C | ↑ (F + C) | [49] |
| Cucumis sativus L. | Chilling | 8 °C | ↑ (F + C) | [50] |
| Hordeum spontaneum L. | Drought | PEG 6000 (−0.75 to −1.5 MPa) | ↑ | [51] |
| Hordeum vulgare L. | Drought | PEG 6000 (−0.5 MPa) | ↑ | [52] |
| Scutellaria baicalensis Georgi | Drought | PEG 6000 (15%) | ↓ (F + T) | [53] |
| Scutellaria baicalensis Georgi | Salt | NaCl (150 mM) | ↑ (F + T) | [53] |
| Hordeum vulgare L. | UV-B radiation | UV-B (0.84 W m−2) | ↑ | [52] |
| Arabidopsis thaliana L. (Col-0) | Iron deficiency | –Fe (0 µM) | ↑ (F) | [54] |
| Gossypium hirsutum L. | Nitrogen deficiency | –N (0 µM) | ↑ | [55] |
| Solanum lycopersicum L. | Alkalinity | pH 9.0 buffer | ↑ | [56] |
SA can also accumulate in roots in response to abiotic stresses such as aluminum, cadmium, chilling, salt, drought, UV-B radiation exposure, alkalinity, and iron- and nitrogen deficiency (Table 1), echoing the reported role of SA in abiotic stress resistance (reviewed in [57][58]). In some cases, stress-induced SA accumulates locally in the root, which comes into direct contact with the stress factor, but it can also be transported to the aboveground tissue. For example, in barley, SA accumulates in response to drought in the roots but not in shoots [51]. In grapes exposed to heat stress, SA is progressively transported from the roots to shoots via xylem [59]. Stress-induced changes in endogenous SA levels are species-specific. For example, drought increases SA content in barley [51] but reduces it in Scutellaria baicalensis roots [53].
SA (50 μM) treatment promotes adventitious root development at the base of cucumber hypocotyls and strongly increases endogenous SA levels in the rooting zone [60]. It is worthy of note that an SA treatment does not necessarily elevate endogenous SA levels in the root due to exogenous SA uptake. For example, priming wheat seed with SA (50 µM, 3 h) or treating 10 DAG seedlings with 500 µM SA for 1–24 h reduces the endogenous levels of both free and conjugated SA in roots [6][47]. The endogenous levels of total and free SA in Scutellaria baicalensis roots also decrease when seedlings are treated with 300 µM SA [53]. Therefore, the type and intensity of exogenous SA effects on plant growth are probably related to changes in the plant’s endogenic SA content and/or redistribution of free and conjugated forms. Accordingly, a feasible role of SA biosynthesis in endogenous SA content after an exogenous SA treatment was demonstrated in several studies [61][62]. Priming maize seeds with [3,4,5,6-2 H4]-salicylic acid (D4SA; the SA deuterated isotopomer) during germination allowed researchers to estimate both SA uptake and SA’s regulation of its own biosynthesis in developing roots [63]. A low SA concentration (50 µM) increased both SA uptake and biosynthesis, whereas a high SA level (500 µM) more strongly enhanced SA uptake but inhibited SA biosynthesis.
Growing evidence indicates that normal plant growth requires that optimal levels of endogenous SA are maintained. Accordingly, a number of negative regulators that alleviate SA accumulation (such as CPR5, DND1, PI4KIIIꞵ1, PI4KIIIꞵ2 etc.) were described in relation to Arabidopsis [7][8][64][65]. Moreover, hybrids between Arabidopsis accessions with suboptimal and supraoptimal SA content (Columbia and C24, respectively) show root growth heterosis [66]. The chromatin remodeler DECREASED DNA METHYLATION 1 (DDM1) links heterosis with endogenous SA levels. Columbia/C24 hybrid heterosis in the root length is impaired in the ddm1 mutant background.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23042228
References
- Raskin, I. Salicylate, A New Plant Hormone. Plant Physiol. 1992, 99, 799–803.
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36.
- Chen, J.; Zhang, J.; Kong, M.; Freeman, A.; Chen, H.; Liu, F. More stories to tell: NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1, a salicylic acid receptor. Plant Cell Environ. 2021, 44, 1716–1727.
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338.
- Chen, Z.; Iyer, S.; Caplan, A.; Klessig, D.F.; Fan, B. Differential accumulation of salicylic acid and salicylic acid-sensitive catalase in different rice tissues. Plant Physiol. 1997, 114, 193–201.
- Rakhmankulova, Z.F.; Fedyaev, V.V.; Rakhmatulina, S.R.; Ivanov, C.P.; Gilvanova, I.R.; Usmanov, I.Y. The effect of wheat seed presowing treatment with salicylic acid on its endogenous content, activities of respiratory pathways, and plant antioxidant status. Russ. J. Plant Physiol. 2010, 57, 778–783.
- Sašek, V.; Janda, M.; Delage, E.; Puyaubert, J.; Guivarc’h, A.; López Maseda, E.; Dobrev, P.I.; Caius, J.; Bóka, K.; Valentová, O.; et al. Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol. 2014, 203, 805–816.
- Janda, M.; Šašek, V.; Ruelland, E. The Arabidopsis pi4kIIIβ1β2 double mutant is salicylic acid-overaccumulating: A new example of salicylic acid influence on plant stature. Plant Signal Behav. 2014, 9, e977210.
- Armengot, L.; Marquès-Bueno, M.M.; Soria-Garcia, A.; Müller, M.; Munné-Bosch, S.; Martínez, M.C. Functional interplay between protein kinase CK 2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J. 2014, 78, 411–423.
- König, S.; Feussner, K.; Schwarz, M.; Kaever, A.; Iven, T.; Landesfeind, M.; Ternes, P.; Karlovsky, P.; Lipka, V.; Feussner, I. Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 2012, 196, 1086–1097.
- Meng, Z.; Ruberti, C.; Gong, Z.; Brandizzi, F. CPR5 modulates salicylic acid and the unfolded protein response to manage tradeoffs between plant growth and stress responses. Plant J. 2017, 89, 486–501.
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 Functions in Salicylic Acid Biosynthesis to Maintain Proper Reactive Oxygen Species Levels for Root Meristem Activity in Rice. Plant Cell 2017, 29, 560–574.
- Kusumi, K.; Yaeno, T.; Kojo, K.; Hirayama, M.; Hirokawa, D.; Yara, A.; Iba, K. The role of salicylic acid in the glutathione-mediated protection against photooxidative stress in rice. Physiol. Plant. 2006, 128, 651–661.
- Stacey, G.; McAlvin, C.B.; Kim, S.Y.; Olivares, J.; Soto, M.J. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiol. 2006, 141, 1473–1481.
- Raskin, I.; Skubatz, H.; Tang, W.; Meeuse, B.J. Salicylic acid levels in thermogenic and non-thermogenic plants. Ann. Bot. 1990, 66, 369–373.
- Mishra, A.K.; Baek, K.H. Salicylic Acid Biosynthesis and Metabolism: A Divergent Pathway for Plants and Bacteria. Biomolecules 2021, 11, 705.
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791.
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565.
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338.
- Hartmann, M.; Zeier, J. N-hydroxypipecolic acid and salicylic acid: A metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 2019, 50, 44–57.
- Li, P.; Cai, Q.; Wang, H.; Li, S.; Cheng, J.; Li, H.; Yu, Q.; Wu, S. Hydrogen peroxide homeostasis provides beneficial micro-environment for SHR-mediated periclinal division in Arabidopsis root. New Phytol. 2020, 228, 1926–1938.
- Takács, Z.; Poór, P.; Borbély, P.; Czékus, Z.; Szalai, G.; Tari, I. H2O2 homeostasis in wild-type and ethylene-insensitive Never ripe tomato in response to salicylic acid treatment in normal photoperiod and in prolonged darkness. Plant Physiol. Biochem. 2018, 126, 74–85.
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856.
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and Extracellular Journey of the Phytohormone Salicylic Acid. Front. Plant Sci. 2019, 10, 423.
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic Acid Affects Root Meristem Patterning via Auxin Distribution in a Concentration-Dependent Manner. Plant Physiol. 2019, 180, 1725–1739.
- Tan, S.; Abas, M.; Verstraeten, I.; Glanc, M.; Molnár, G.; Hajný, J.; Lasák, P.; Petřík, I.; Russinova, E.; Petrášek, J.; et al. Salicylic Acid Targets Protein Phosphatase 2A to Attenuate Growth in Plants. Curr. Biol. 2020, 30, 381–395.
- Klessig, D.F.; Tian, M.; Choi, H.W. Multiple Targets of Salicylic Acid and Its Derivatives in Plants and Animals. Front. Immunol. 2016, 7, 206.
- Slaymaker, D.H.; Navarre, D.A.; Clark, D.; Del Pozo, O.; Martin, G.B.; Klessig, D.F. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 11640–11645.
- Choi, H.W.; Manohar, M.; Manosalva, P.; Tian, M.; Moreau, M.; Klessig, D.F. Activation of Plant Innate Immunity by Extracellular High Mobility Group Box 3 and Its Inhibition by Salicylic Acid. PLoS Pathog. 2016, 12, e1005518.
- Manohar, M.; Wang, D.; Manosalva, P.M.; Choi, H.W.; Kombrink, E.; Klessig, D.F. Members of the abscisic acid co-receptor PP2C protein family mediate salicylic acid-abscisic acid crosstalk. Plant Direct. 2017, 1, e00020.
- Innes, R. The Positives and Negatives of NPR: A Unifying Model for Salicylic Acid Signaling in Plants. Cell 2018, 173, 1314–1315.
- Pokotylo, I.; Kravets, V.; Ruelland, E. Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. Int. J. Mol. Sci. 2019, 20, 4377.
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225.
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Chen, Y.; Wenig, M.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250.
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864.
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101.
- Segarra, G.; Jáuregui, O.; Casanova, E.; Trillas, I. Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 2006, 67, 395–401.
- Branch, C.; Hwang, C.F.; Navarre, D.A.; Williamson, V.M. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol. Plant Microbe Interact. 2004, 17, 351–356.
- Wubben, M.J.E.; Jin, J.; Baum, T.J. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol. Plant Microbe Interact. 2008, 21, 424–432.
- De Vleesschauwer, D.; Van Buyten, E.; Satoh, K.; Balidion, J.; Mauleon, R.; Choi, I.R.; Vera-Cruz, C.; Kikuchi, S.; Höfte, M. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 2012, 158, 1833–1846.
- Denancé, N.; Ranocha, P.; Oria, N.; Barlet, X.; Rivière, M.P.; Yadeta, K.A.; Hoffmann, L.; Perreau, F.; Clément, G.; Maia-Grondard, A.; et al. Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 2013, 73, 225–239.
- Agtuca, B.; Rieger, E.; Hilger, K.; Song, L.; Robert, C.A.; Erb, M.; Karve, A.; Ferrieri, R.A. Carbon-11 reveals opposing roles of auxin and salicylic acid in regulating leaf physiology, leaf metabolism, and resource allocation patterns that impact root growth in Zea mays. J. Plant Growth Regul. 2014, 33, 328–339.
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168.
- Yang, Z.M.; Wang, J.; Wang, S.H.; Xu, L.L. Salicylic acid-induced aluminum tolerance by modulation of citrate efflux from roots of Cassia tora L. Planta 2003, 217, 168–174.
- Liu, N.; Song, F.; Zhu, X.; You, J.; Yang, Z.; Li, X. Salicylic acid alleviates aluminum toxicity in soybean roots through modulation of reactive oxygen species metabolism. Front. Chem. 2017, 5, 96.
- Metwally, A.; Finkemeier, I.; Georgi, M.; Dietz, K.J. Salicylic acid alleviates the cadmium toxicity in barley seedlings. Plant Physiol. 2003, 132, 272–2781.
- Tajti, J.; Németh, E.; Glatz, G.; Janda, T.; Pál, M. Pattern of changes in salicylic acid-induced protein kinase (SIPK) gene expression and salicylic acid accumulation in wheat under cadmium exposure. Plant Biol. 2019, 21, 1176–1180.
- Zhao, Q.; Gu, C.; Sun, Y.; Li, G.; Li, L.L.; Hao, L. Root defense in salicylic acid-altering Arabidopsis plants in responses to cadmium stress. J. Plant Growth Regul. 2021, 40, 1764–1776.
- Wang, D.H.; Li, X.X.; Su, Z.K.; Ren, H.X. The role of salicylic acid in response of two rice cultivars to chilling stress. Biol. Plant. 2009, 53, 545.
- Dong, C.J.; Li, L.; Shang, Q.M.; Liu, X.Y.; Zhang, Z.G. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 2014, 240, 687–700.
- Bandurska, H.; Stroiński, A. The effect of salicylic acid on barley response to water deficit. Acta Physiol. Plant. 2005, 27, 379–386.
- Bandurska, H.; Cieślak, M. The interactive effect of water deficit and UV-B radiation on salicylic acid accumulation in barley roots and leaves. Environ. Exp. Bot. 2013, 94, 9–18.
- Su, H.; Song, S.; Yan, X.; Fang, L.; Zeng, B.; Zhu, Y. Endogenous salicylic acid shows different correlation with baicalin and baicalein in the medicinal plant Scutellaria baicalensis Georgi subjected to stress and exogenous salicylic acid. PLoS ONE 2018, 13, e0192114.
- Shen, C.; Yang, Y.; Liu, K.; Zhang, L.; Guo, H.; Sun, T.; Wang, H. Involvement of endogenous salicylic acid in iron-deficiency responses in Arabidopsis. J. Exp. Bot. 2016, 67, 4179–4193.
- Chen, J.; Liu, S.; Zhang, S.; Ge, C.; Shen, Q.; Ma, H.; Zhang, X.; Dong, H.; Zhao, X.; Pang, C. Nitrogen modulates cotton root morphology by affecting abscisic acid (ABA) and salicylic acid (SA) content. Arch. Agron. Soil Sci. 2020, 67, 1722–1738.
- Khan, A.; Kamran, M.; Imran, M.; Al-Harrasi, A.; Al-Rawahi, A.; Al-Amri, I.; Lee, I.J.; Khan, A.L. Silicon and salicylic acid confer high-pH stress tolerance in tomato seedlings. Sci. Rep. 2019, 9, 1–16.
- Guo, B.; Liu, C.; Liang, Y.; Li, N.; Fu, Q. Salicylic Acid Signals Plant Defence against Cadmium Toxicity. Int, J. Mol. Sci. 2019, 20, 2960.
- Khan, M.I.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462.
- Liu, H.T.; Liu, Y.P.; Huang, W.D. Root-fed salicylic acid in grape involves the response caused by aboveground high temperature. J. Integr. Plant Biol. 2008, 50, 761–767.
- Dong, C.J.; Liu, X.Y.; Xie, L.L.; Wang, L.L.; Shang, Q.M. Salicylic acid regulates adventitious root formation via competitive inhibition of the auxin conjugation enzyme CsGH3. 5 in cucumber hypocotyls. Planta 2020, 252, 1–15.
- Szalai, G.; Horgosi, S.; Soós, V.; Majláth, I.; Balázs, E.; Janda, T. Salicylic acid treatment of pea seeds induces its de novo synthesis. J. Plant Physiol. 2011, 168, 213–219.
- Huang, Y.T.; Cai, S.Y.; Ruan, X.L.; Chen, S.Y.; Mei, G.F.; Ruan, G.H.; Cao, D.D. Salicylic acid enhances sunflower seed germination under Zn2+ stress via involvement in Zn2+ metabolic balance and phytohormone interactions. Sci. Hortic. 2021, 275, 109702.
- Moravcová, Š.; Tůma, J.; Dučaiová, Z.K.; Waligórski, P.; Kula, M.; Saja, D.; Słomka, A.; Bąba, W.; Libik-Konieczny, M. Influence of salicylic acid pretreatment on seeds germination and some defence mechanisms of Zea mays plants under copper stress. Plant Physiol. Biochem. 2018, 122, 19–30.
- Bowling, S.A.; Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 1997, 9, 1573–1584.
- Clough, S.J.; Fengler, K.A.; Yu, I.C.; Lippok, B.; Smith, R.K., Jr.; Bent, A.F. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 2000, 97, 9323–9328.
- Zhang, Q.; Li, Y.; Xu, T.; Srivastava, A.K.; Wang, D.; Zeng, L.; Yang, L.; He, L.; Zhang, H.; Zheng, Z.; et al. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2016, 2, 6027.
This entry is offline, you can click here to edit this entry!
