Diagnostic tools play a pivotal role in warfare against schistosomiasis but must adapt to the endemic status and objectives of activities. With the decrease of prevalence and infection intensity of schistosomiasis in human beings and livestock, optimal methodologies with high sensitivity and absolute specificity are needed for the detection of asymptomatic cases or light infections, as well as disease surveillance to verify elimination. In comparison with the parasitological methods with relatively low sensitivity and serological techniques lacking specificity, which both had been widely used in previous control stages, the molecular detection methods based on the amplification of promising genes of the schistosome genome may pick up the baton to assist the eventual aim of elimination.
- schistosomiasis japonica
- elimination
- diagnostic tools
- molecular techniques
1. Traditional Diagnostic Tools Applied in Schistosomiasis Control in China
It cannot be exaggerated that diagnosis is the essential basis of schistosomiasis control for case identification and treatment, assessment of morbidity, and evaluation of control strategies, which are all dependent on the performance of diagnostic tests [27,39]. Two kinds of diagnostic methodologies, namely parasitological techniques mainly including KK and MHT [40,41], and immunologic approaches based on detection of specific antibodies, were widely used in the national control program in China, accelerating the process of schistosomiasis control significantly [11,42].
1.1. Parasitological Methods
1.2. Immunologic Tests
2. Molecular Methods Developed to Detect the Pathogen of Schistosomiasis
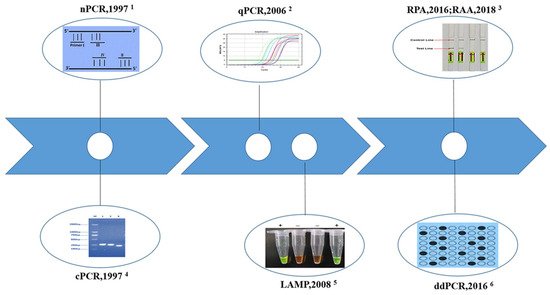
2.1. Conventional Polymerase Chain Reaction (cPCR)
2.2. Nested PCR (nPCR)
2.3. Real-Time Quantitative PCR (qPCR)
2.4. Droplet Digital PCR (ddPCR)
2.5. Loop-Mediated Isothermal Amplification (LAMP)
2.6. Recombinase Polymerase Amplification (RPA)
This entry is adapted from the peer-reviewed paper 10.3390/pathogens11030287
