The bacterium Proteus terrae is a non-pathogenic and natural microflora of humans, but Morganella morganii is an opportunistic pathogen. These organisms belong to risk group II. Screening the sensitivity of these isolates to antibiotics revealed a sufficient number of antibiotics, which can be used to control them, if required. BR3 and BR4 exhibited resistance to individual antibiotics, ampicillin and vancomycin, whereas only BR3 was resistant to tetracycline. The current investigation, along with earlier reported work on these isolates, identifies BR3 as a useful isolate in the industrial application for the degradation of EtBr. Identical and related microorganisms, which are available in the culture collection repositories, can also be explored for such potential to formulate a microbial consortium for the bioremediation of ethidium bromide prior to its disposal.
- bioaccumulation
- risk assessment
- inhibition zone
- biotransformation
- 16S-rRNA
- Proteus terrae
- Morganella morganii
- pathogenicity
1. Introduction
2. Current Insights
2.1. Antibiotic Resistance Assay and Gel Electrophoresis of Crude Lysate
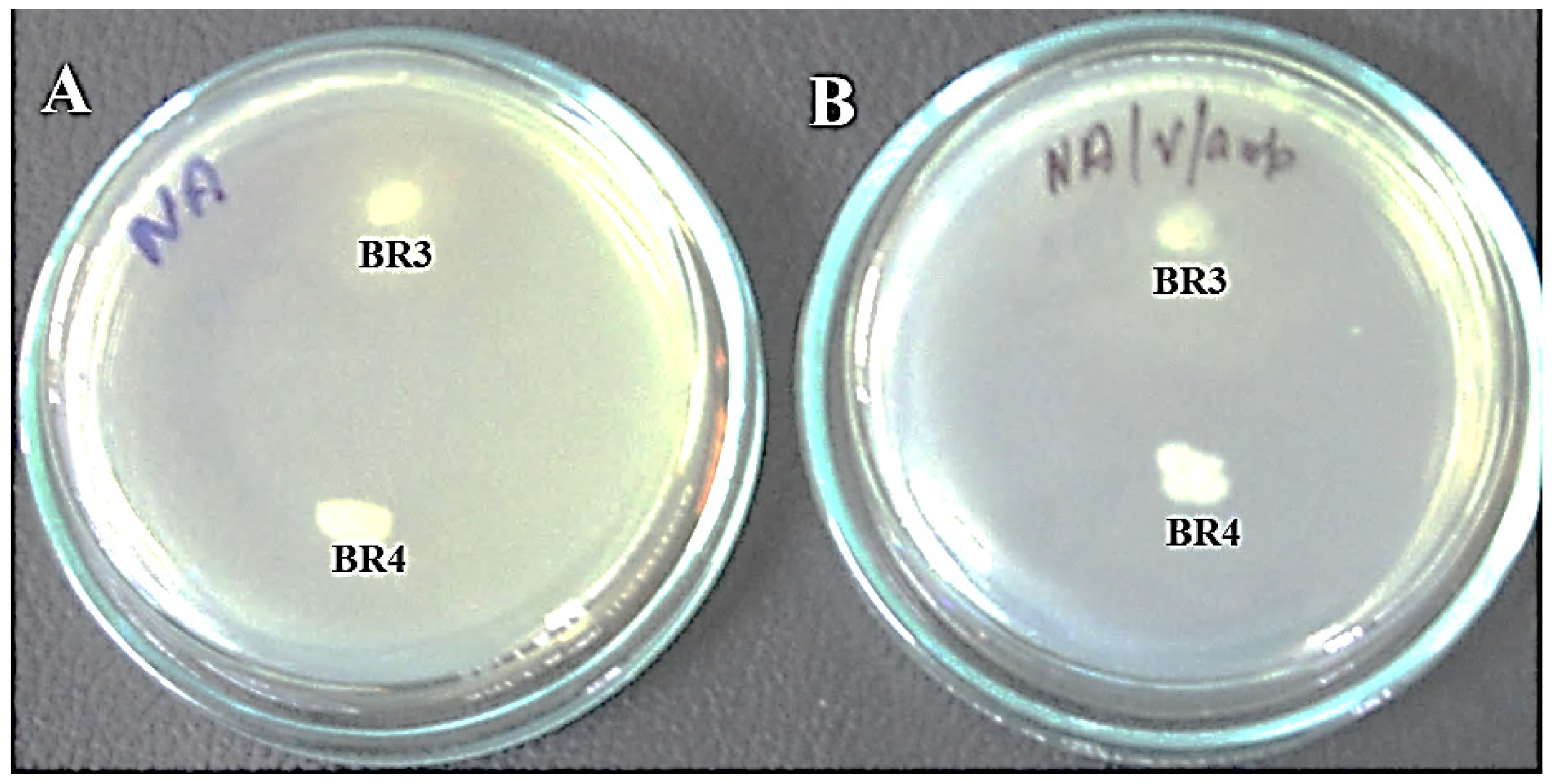 Figure 1. Growth of isolates BR3 and BR4 on nutrient agar plates. (A) Nutrient agar plate without ampicillin. (B) Nutrient agar plate with ampicillin.
Figure 1. Growth of isolates BR3 and BR4 on nutrient agar plates. (A) Nutrient agar plate without ampicillin. (B) Nutrient agar plate with ampicillin.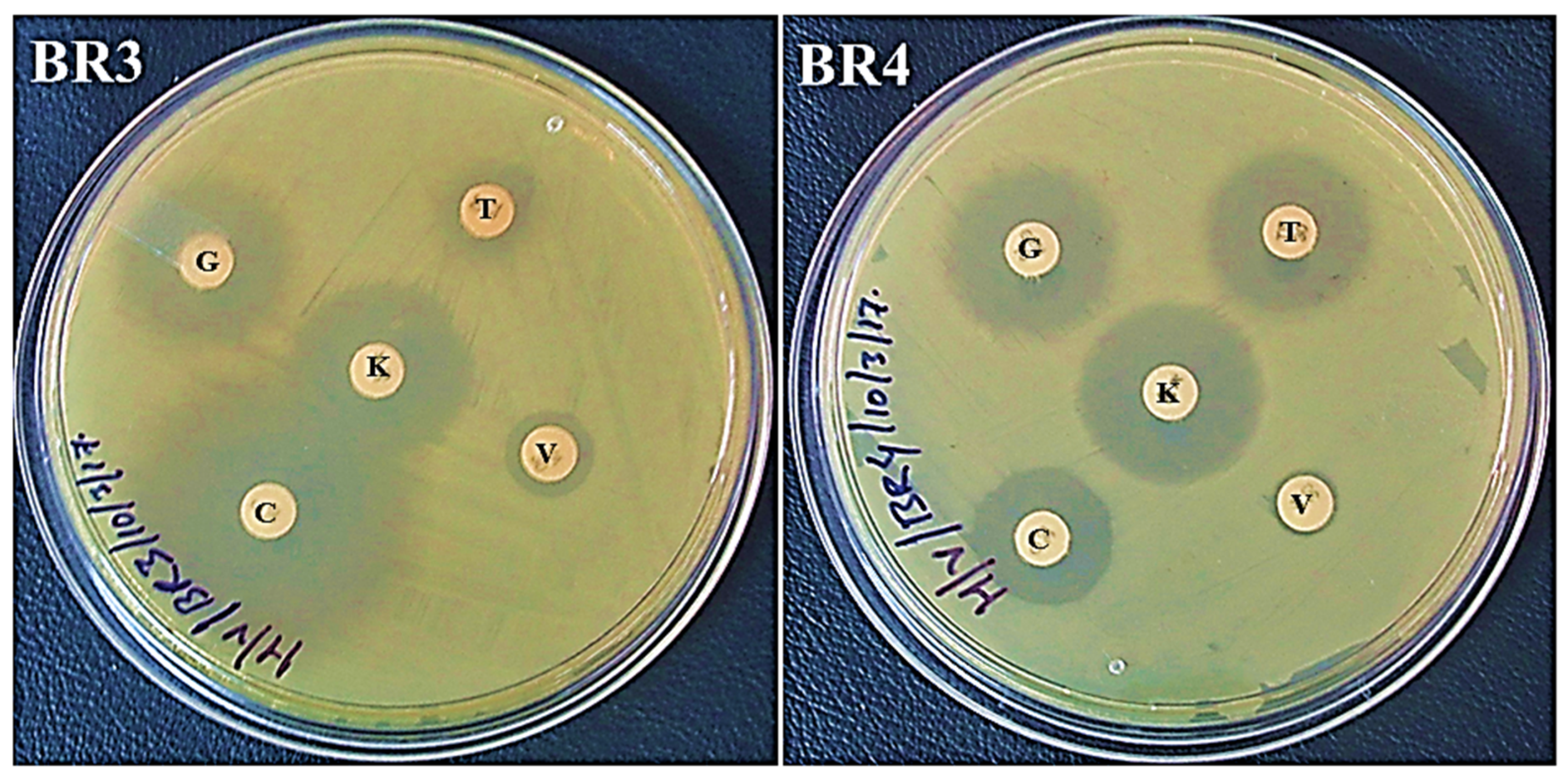 Figure 2. Antibiotic sensitivity profile of BR3 and BR4 on Mueller–Hinton agar plates. Letters on the discs represent antibiotics: C: chloramphenicol (30 µg/disc), V: vancomycin (30 µg/disc), T: tetracycline (30 µg/disc), K: kanamycin (30 µg/disc), and G: gentamicin (10 µg/disc). The inner diameter of the medium holding plate measures 86 mm.
Figure 2. Antibiotic sensitivity profile of BR3 and BR4 on Mueller–Hinton agar plates. Letters on the discs represent antibiotics: C: chloramphenicol (30 µg/disc), V: vancomycin (30 µg/disc), T: tetracycline (30 µg/disc), K: kanamycin (30 µg/disc), and G: gentamicin (10 µg/disc). The inner diameter of the medium holding plate measures 86 mm.| Diameter (mm) of the Zone of Inhibition Following 24 h of Incubation | |||||
|---|---|---|---|---|---|
| Bacterial Isolates | Antibiotics | ||||
| C | V | T | K | G | |
| BR3 | 32 (S) | 11 (R) | 11 (R) | 23 (S) | 22 (S) |
| BR4 | 16 (I) | <1 (R) | 20 (S) | 22 (S) | 20 (S) |
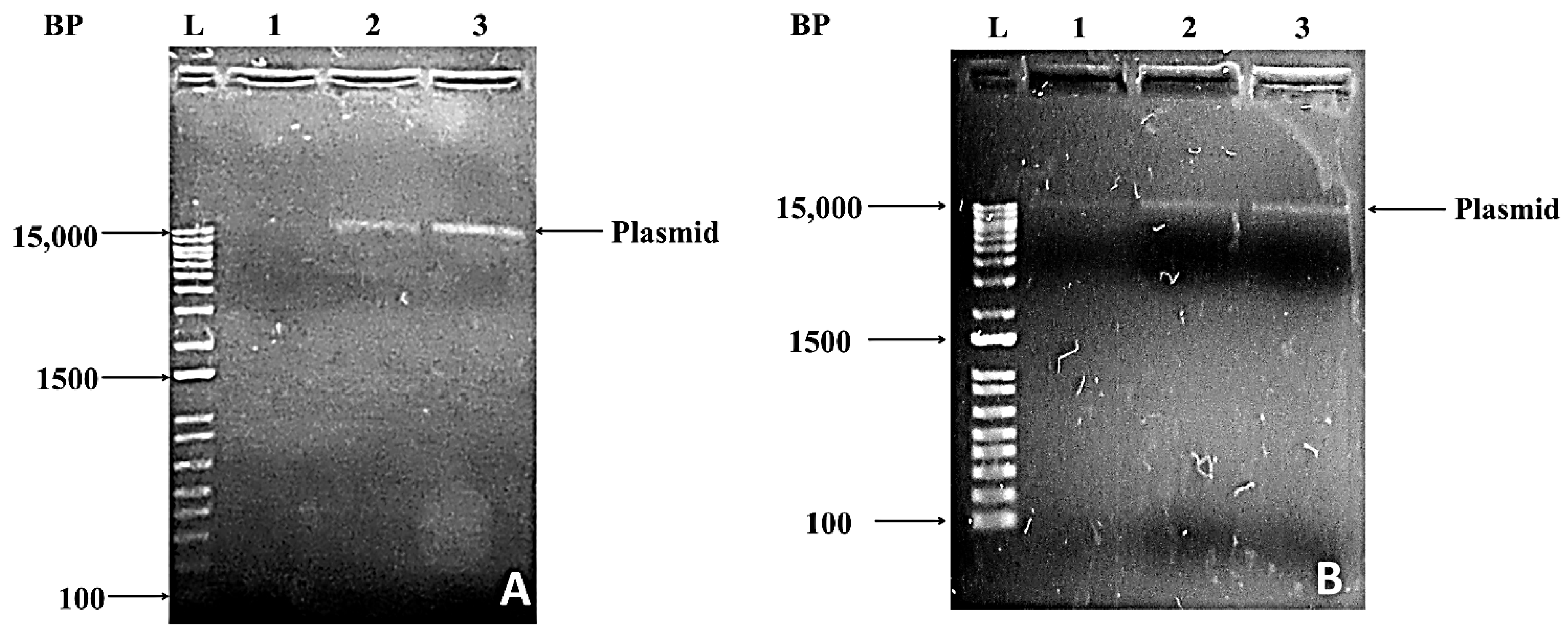 Figure 3. A total of 1.2% agarose gel electrophoresis of plasmid preparation from bacterial isolates BR3 (A) and BR4 (B). Lanes 1–3 represent the plasmid preparation from culture grown in 1: LB medium, 2: LB medium supplemented with ethidium bromide, and 3: LB medium supplemented with ampicillin. L represents the molecular weight marker; the molecular weight ranges from 100–15,000 bp.
Figure 3. A total of 1.2% agarose gel electrophoresis of plasmid preparation from bacterial isolates BR3 (A) and BR4 (B). Lanes 1–3 represent the plasmid preparation from culture grown in 1: LB medium, 2: LB medium supplemented with ethidium bromide, and 3: LB medium supplemented with ampicillin. L represents the molecular weight marker; the molecular weight ranges from 100–15,000 bp.2.2. Identification and Phylogenetic Status of Bacterial Isolates
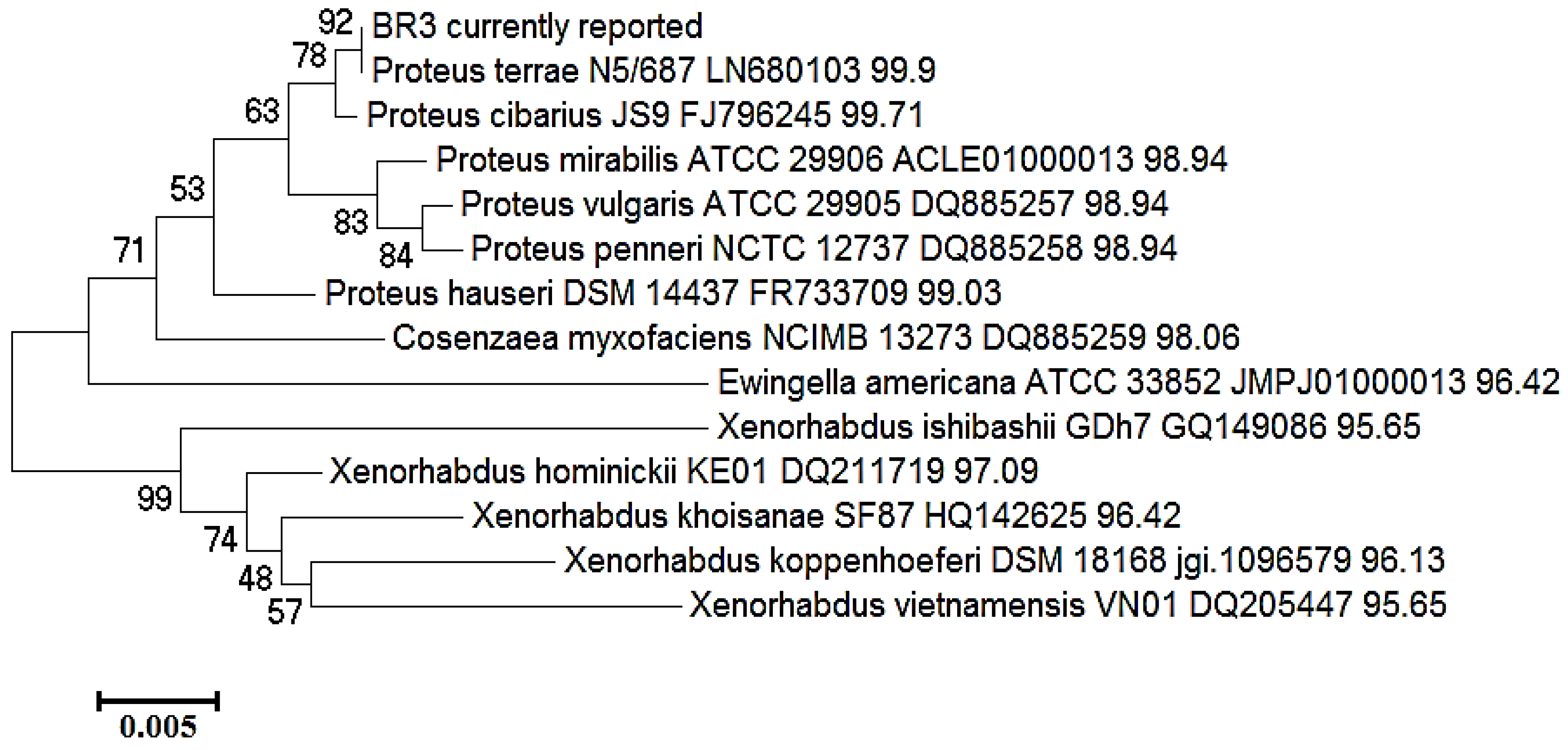 Figure 4. Phylogenetic tree based on 16S-rRNA showing the clustering of BR3 with sequences retrieved form the EzBioCloud database. The tree was constructed in MEGAX by the neighbor-joining method with a 500-bootstrap value.
Figure 4. Phylogenetic tree based on 16S-rRNA showing the clustering of BR3 with sequences retrieved form the EzBioCloud database. The tree was constructed in MEGAX by the neighbor-joining method with a 500-bootstrap value.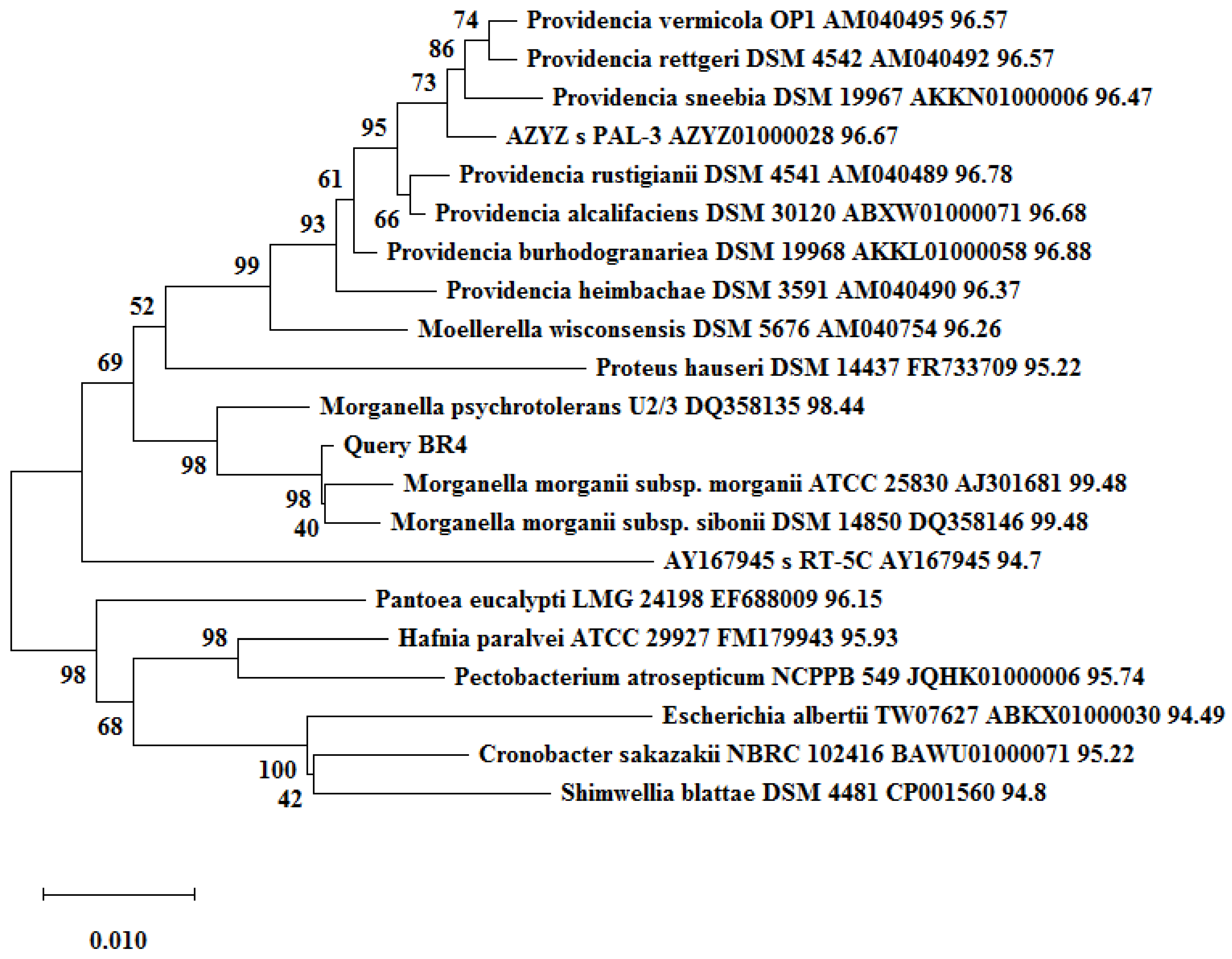 Figure 5. Phylogenetic tree based on 16S-rRNA showing the clustering of BR4 with sequences retrieved form the EzBioCloud database. The tree was constructed in MEGAX by the neighbor-joining method with a 500 bootstrap value.
Figure 5. Phylogenetic tree based on 16S-rRNA showing the clustering of BR4 with sequences retrieved form the EzBioCloud database. The tree was constructed in MEGAX by the neighbor-joining method with a 500 bootstrap value.2.3. Pathogenicity Status of the Isolates
This entry is adapted from the peer-reviewed paper 10.3390/biotech11010004
References
- Lunn, G.; Sansone, E.B. Ethidium bromide: Destruction and decontamination of solutions. Anal. Biochem. 1987, 162, 453–458.
- Zocher, R.; Billich, A.; Keller, U.; Messner, P. Destruction of ethidium bromide in solution by ozonolysis. Biol. Chem. Hoppe-Seyler 1988, 369, 1191–1194.
- Armour, M.A. Hazardous Laboratory Chemicals Disposal Guide, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2003; pp. 256–257.
- Joshua, H. Quantitative adsorption of ethidium-bromide from aqueous-solutions by macroreticular resins. Biotechniques 1986, 4, 207–208. Available online: https://Biotechniques.org/Quantitative-adsorption-of-ethidium-bromide-from-aqueous-solutions-by-macroreticular-resins (accessed on 28 July 2020).
- Sigma-Aldrich and Merck. Ethidium Bromide—Product Information Sheet. Available online: https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/e7637pis.pdf (accessed on 29 July 2020).
- Xie, E.; Zheng, L.; Ding, A.; Zhang, D. Mechanisms and pathways of ethidium bromide Fenton-like degradation by reusable magnetic nanocatalysts. Chemosphere 2021, 262, 127852.
- Sukhumungoon, P.; Rattanachuay, P.; Hayeebilan, F.; Kantachote, D. Biodegradation of ethidium bromide by Bacillus thuringiensis isolated from soil. Afr. J. Microbiol. Res. 2013, 7, 471–476. Available online: https://academicjournals.org/journal/AJMR/article-full-text-pdf/6F49CBE19946 (accessed on 28 July 2020).
- Lone, T.A.; Revathi, C.; Lone, R.A. Isolation of dye degrading Bacillus species from the soil near dyeing industry and its potential application in dye effluent treatment. Am.-Eurasian J. Toxicol. Sci. 2015, 7, 129–135.
- Patil, S.M.; Berde, C.V. Biodegradation of carcinogenic dye, ethidium bromide, by soil microorganisms. World J. Pharm. Pharm. Sci. 2015, 4, 1210–1219. Available online: https://storage.googleapis.com/journal-uploads/wjpps/article_issue/1435661544.pdf. (accessed on 28 July 2020).
- Kumar, A.; Swarupa, P.; Gandhi, V.P.; Kumari, S. Isolation of Ethidium Bromide Degrading Bacteria from Jharkhand. Int. J Appl. Sci. Biotechnol. 2017, 5, 293–301.
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. Principles of Fermentation Technology, 2nd ed.; Elsevier Sciences: Oxford, UK, 2013.
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17.
- Razalski, A.; Sidorczyk, Z.; Kotea, K. Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 1997, 61, 65–89. Available online: https://Microbiologyandmolecularbiologyreview.org/Potential-virulence-factors-of-Proteus-bacilli (accessed on 25 July 2020).
- Wang, H.F.; Du, L.Y.; Luo, J.; He, H.X. Isolation, identification and characterization of Morganella morganii from Naja naja atra in Beijing, China. Cell. Mol. Biol. 2017, 63, 52–58.
- Dai, H.; Lu, B.; Li, Z.; Huang, Z.; Cai, H.; Yu, K.; Wang, D. Multilocus sequence analysis for the taxonomic updating and identification of the genus Proteus and reclassification of Proteus genospecies 5 O’Hara et al. 2000, Proteus cibarius Hyun et al. 2016 as later heterotypic synonyms of Proteus terrae Behrendt et al. 2015. BMC Microbiol. 2020, 20, 152.
- Classification of Microorganisms by Risk Group, UNSW, Australia. Available online: https://safety.unsw.edu.au/sites/default/files/HS076_Classification_of_infective_microorganisms.pdf (accessed on 29 July 2020).
- Wayne, P.A. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement, CLSI Document M100-S25; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2015.
- Chung, T.H.; Yi, S.W.; Kim, B.S.; Kim, W.I.; Shin, G.W. Identification and antibiotic resistance profiling of bacterial isolates from septicaemic soft-shelled turtles (Pelodiscus sinensis). Veterinární Med. 2017, 62, 169–177.
- Behrendt, U.; Augustin, J.R.; Spraer, C.; Gelbrecht, J.; Schumann, P.; Ulrich, A. Taxonomic characterisation of Proteus terrae sp. nov., a N2O-producing, nitrate-ammonifying soil bacterium. Antonie Leeuwenhoek 2015, 108, 1457–1468.
- Yu, X.; Torzewska, A.; Zhang, X.; Yin, Z.; Drzewiecka, D.; Cao, H.; Liu, B.; Knirel, Y.A.; Rozalski, A.; Wang, L. Genetic diversity of the O antigens of Proteus species and the development of a suspension array for molecular serotyping. PLoS ONE 2017, 12, e0183267.
