Cereals are an important and major source of the human diet. They constitute more than two-thirds of the world’s food source and cover more than 56% of the world’s cultivatable land. These important sources of food are affected by a variety of damaging diseases, causing significant loss in annual production. In this regard, detection of diseases at an early stage and quantification of the severity has acquired the urgent attention of researchers worldwide. One emerging and popular approach for this task is the utilization of machine learning techniques.
- cereal crop
- plant disease
- machine learning
- deep learning
1. Introduction
2. Cereal Crops and Diseases
2.1. Wheat
|
Pathogen |
Disease |
|---|---|
|
Leaf Rust (Brown Rust), Stem Rust (Black Rust), |
|
|
Stripe Rust (Yellow Rust), Common Root Rot, |
|
|
Fungus |
Common and Dwarf Bunt (Stinking Smut), |
|
Wheat Blast, |
|
|
Tan Spot |
|
|
Bacterial Stripe (Black Chaff), |
|
|
Bacteria |
Basal Glume Rot and Bacterial Leaf Blight, |
|
Bacterial Spike Blight (Gummosis) |
|
|
Barley Yellow Dwarf, |
|
|
Virus |
Barley Stripe Mosaic, |
|
Wheat Streak Mosaic |
|
|
Aphids, Stink Bugs, |
|
|
Cereail Leaf Beetle, |
|
|
Insect |
Thrips, |
|
Hessian Fly, Wireworms, |
|
|
Mites |
|
|
Nematode |
Seed Gall Nematode |
|
Cereal Cyst Nematode |
|
|
Root Knot Nematode |
|
|
Root Lesion Nematode |
2.2. Maize (Corn)
|
Pathogen |
Disease |
|---|---|
|
Fungus |
Gray leaf spot, Brown spot, |
|
Stripe Rust (Yellow Rust) |
|
|
Common rust, Smut, |
|
|
Northernl eaf blight, Southern leaf blight |
|
|
Bacteria |
Corn stunt disease |
|
Stewart wilt |
|
|
Bacterial stalk rot |
|
|
Bacterial leaf strip |
|
|
Virus |
Leaf fleek |
|
Mosaic |
|
|
Yellow dwarf |
2.3. Rice
2.4. Barley
2.5. Sorghum
|
Pathogen |
Disease |
|---|---|
|
Fungus |
Anthracnose, Leaf blight, Zonate leaf spot |
|
Tar spot, Charcoal rot |
|
|
Rust, Gray leaf spot |
|
|
Bacteria |
Bacterial stripe |
|
Virus |
Streak disease |
|
Pathogen |
Disease |
|---|---|
|
Fungus |
Crown rust, Stem rust, Powdery mildew |
|
Smut disease, Leaf blight |
|
|
Root rot, Crown rot, Snow mold |
|
|
Bacteria |
Halo blight |
|
Virus |
Yellow dwarf |
|
Mosaic |
|
|
golden stripe |
|
Pathogen |
Disease |
|---|---|
|
Fungus |
Snow mold, Brown rust, Ergot |
|
Eye spot, Sharp eyespot |
|
|
Powdery mildew, Stem rust, Glume blotch |
|
|
Virus |
Yellow dwarf |
3. Machine Learning-Based Cereal Crop Disease Detection
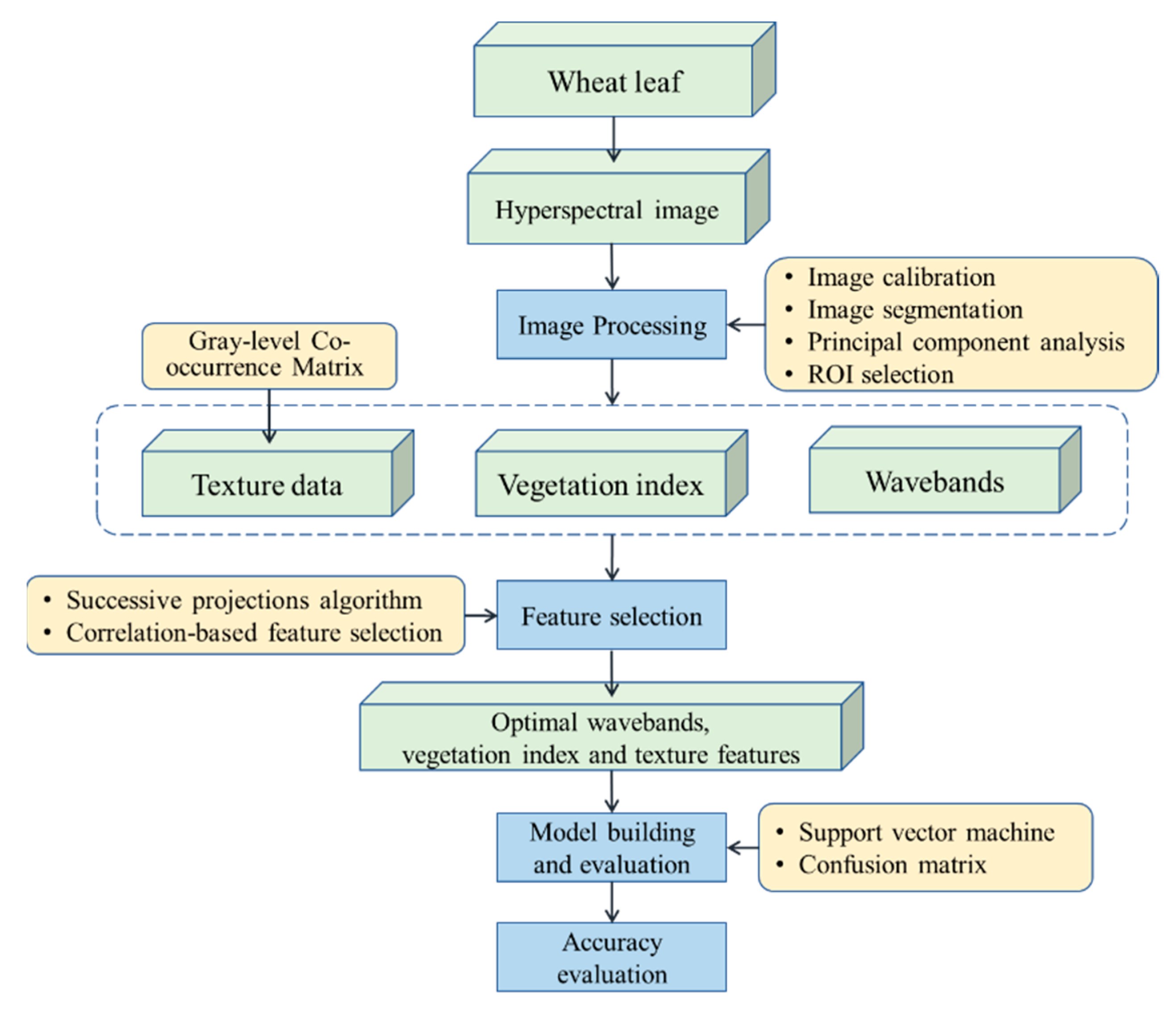
3.1. Machine Learning in Wheat Disease Detection

|
Citatation |
Year |
Data Type |
# of Classes |
Sample Size |
Method |
Accuracy % |
|---|---|---|---|---|---|---|
|
Bao et al. [38] |
2021 |
Image |
3 |
360 |
SVM |
93.3% |
|
Sood et al. [45] |
2020 |
Image |
3 |
876 |
VGG16 |
99.07% |
|
Mukhtar et al. [46] |
2021 |
Image |
11 |
440 |
MobileNet |
92% |
|
Kumar et.al [47] |
2021 |
Image |
1 |
450 |
CNN |
89.9% |
|
Tagel et al. [48] |
2021 |
Image |
3 |
1500 |
VGG19 |
99.38% |
|
Hussain et al. [49] |
2018 |
Image |
4 |
8828 |
AlexNet |
84.54% |
|
Jiang et al. [50] |
2017 |
Image |
6 |
9230 |
VGG-FCN |
97.95% |
|
Azadbakht et al. [51] |
2019 |
Hyper-spectral |
2 |
284 |
v-SVR |
0.99R2 |
|
Huang et al. [40] |
2019 |
Hyper-spectral |
2 |
145 |
Linear Regression |
0.75R2 |
|
Huang et al. [41] |
2019 |
Hyper-spectral |
2 |
89 |
SVM |
85.7% |
3.2. Machine Learning in Rice Disease Detection
|
Citatation |
Year |
Data Type |
# of Classes |
Sample Size |
Method |
Accuracy % |
|---|---|---|---|---|---|---|
|
Chen et al. [52] |
2021 |
Image |
12 |
1100 |
MobileNetV2 |
99.67% |
|
Wang et al. [53] |
2021 |
Image |
3 |
2370 |
MobileNetV2 |
94.65% |
|
Liang et al. [54] |
2019 |
Image |
1 |
5808 |
CNN |
95.83% |
|
Rahman et.al [55] |
2021 |
Image |
3 |
300 |
CNN |
90% |
|
Saha and Ahsan. [57] |
2021 |
Image |
3 |
276 |
CNN |
91.47% |
|
Chen et al. [58] |
2020 |
Image |
15 |
500 |
DenseNet |
94.07% |
|
kamrul et al. [59] |
2019 |
Image |
2 |
284 |
InceptionV3 |
99% |
|
Hasan et al. [60] |
2019 |
Image |
9 |
1080 |
InceptionV3 |
97.5% |
|
Sethy et al. [61] |
2020 |
Image |
11 |
5932 |
SVM |
98.38% |
|
Zhou et al. [62] |
2019 |
Image |
3 |
7448 |
faster R-CNN |
96.21% |
3.3. Machine Learning in Maize Disease Detection
|
Citatation |
Year |
Data Type |
# of Classes |
Sample Size |
Method |
Accuracy % |
|---|---|---|---|---|---|---|
|
Agarwal et al. [52] |
2021 |
Image |
9 |
500 |
CNN |
95.12% |
|
Sibiya et al. [64] |
2019 |
Image (PlantVillage) |
9 |
2500 |
CNN |
95.5% |
|
Barman et al. [65] |
2021 |
Image (PlantVillage) |
9 |
2500 |
MobileNetV2 |
93.5% |
|
Hasan et al. [66] |
2020 |
Image (PlantVillage) |
9 |
2500 |
LSTM |
99.02% |
|
Xu et al. [67] |
2021 |
Image (PlantVillage) |
9 |
2500 |
TCI-ALEXN |
99.18% |
|
Tian [68] |
2019 |
Image (PlantVillage) |
9 |
2500 |
VGG16 |
96.8% |
This entry is adapted from the peer-reviewed paper 10.3390/a15030075
References
- Debelee, T.G.; Kebede, S.R.; Schwenker, F.; Shewarega, Z.M. Deep Learning in Selected Cancers’ Image Analysis—A Survey. J. Imaging 2020, 6, 121.
- Rufo, D.D.; Debelee, T.G.; Ibenthal, A.; Gachena, W.G. Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM). Diagnostics 2021, 11, 1714.
- Debelee, T.G.; Amirian, M.; Ibenthal, A.; Palm, G.; Schwenker, F. Classification of mammograms using convolutional neural network based feature extraction. In Proceedings of the International Conference on Information and Communication Technology for Develoment for Africa, Bahir Dar, Ethiopia, 25–27 September 2017; Springer: Cham, Switzerland, 2017; pp. 89–98.
- Rufo, D.D.; Debelee, T.G.; Gachena, W.G. A Hybrid Machine Learning Model Based on Global and Local Learner Algorithms for Diabetes Mellitus Prediction. J. Biomim. Biomater. Biomed. Eng. 2022, 54, 65–88.
- Biratu, E.S.S.; Schwenker, F.; Debelee, T.G.G.; Kebede, S.R.R.; Negera, W.G.G.; Molla, H.T.T. Enhanced Region Growing for Brain Tumor MR Image Segmentation. J. Imaging 2021, 7, 22.
- Debelee, T.G.; Schwenker, F.; Ibenthal, A.; Yohannes, D. Survey of deep learning in breast cancer image analysis. Evol. Syst. 2020, 11, 143–163.
- Debelee, T.G.; Gebreselasie, A.; Schwenker, F.; Amirian, M.; Yohannes, D. Classification of mammograms using texture and cnn based extracted features. J. Biomim. Biomater. Biomed. Eng. 2019, 42, 79–97.
- Rahimeto, S.; Debelee, T.G.; Yohannes, D.; Schwenker, F. Automatic pectoral muscle removal in mammograms. Evol. Syst. 2019, 12, 519–526.
- Kebede, S.R.; Debelee, T.G.; Schwenker, F.; Yohannes, D. Classifier Based Breast Cancer Segmentation. J. Biomim. Biomater. Biomed. Eng. 2020, 47, 41–61.
- Gelana, F.; Yadav, A. Firearm detection from surveillance cameras using image processing and machine learning techniques. In Smart Innovations in Communication and Computational Sciences; Springer: Singapore, 2019; pp. 25–34.
- Debelee, T.G.; Schwenker, F.; Kebede, S.R.; Yohannes, D. Evaluation of modified adaptive k-means segmentation algorithm. Comput. Vis. Media 2019, 5, 347–361.
- Waldamichael, F.G.; Debelee, T.G.; Ayano, Y.M. Coffee disease detection using a robust HSV color-based segmentation and transfer learning for use on smartphones. Int. J. Intell. Syst. 2011.
- Kibru, Y.; Debelee, T. Detection of Bacterial Wilt on Enset Crop Using Deep Learning Approach. Int. J. Eng. Res. Afr. 2020, 51, 131–146.
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69.
- Nturambirwe, J.F.I.; Opara, U.L. Machine learning applications to non-destructive defect detection in horticultural products. Biosyst. Eng. 2020, 189, 60–83.
- Rasti, S.; Bleakley, C.J.; Holden, N.; Whetton, R.; Langton, D.; O’Hare, G. A Survey of High Resolution Image Processing Techniques for Cereal Crop Growth Monitoring. Inf. Process. Agric. 2021.
- Sharma, R.; Kamble, S.S.; Gunasekaran, A.; Kumar, V.; Kumar, A. A systematic literature review on machine learning applications for sustainable agriculture supply chain performance. Comput. Oper. Res. 2020, 119, 104926.
- Virnodkar, S.S.; Pachghare, V.K.; Patil, V.; Jha, S.K. Remote sensing and machine learning for crop water stress determination in various crops: A critical review. Precis. Agric. 2020, 21, 1121–1155.
- Stubbs, R.; Prescott, J.; Saari, E.; Dubin, H. Cereal Disease Methodology Manual. 1986. Available online: http://hdl.handle.net/10883/3997 (accessed on 21 February 2022).
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116.
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The Most Important Fungal Diseases of Cereals—Problems and Possible Solutions. Agronomy 2021, 11, 714.
- Schaad, N.W.; Frederick, R.D.; Shaw, J.; Schneider, W.L.; Hickson, R.; Petrillo, M.D.; Luster, D.G. Advances in molecular-based diagnostics in meeting crop biosecurity and phytosanitary issues. Annu. Rev. Phytopathol. 2003, 41, 305–324.
- Barton, S. CEREALS | Grain Defects. In Encyclopedia of Grain Science; Wrigley, C., Ed.; Elsevier: Oxford, UK, 2004; pp. 223–228.
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553.
- Duveiller, E.; Singh, R.; Singh, P.; Dababat, A.; Mezzalama, M. Wheat Diseases and Pests: A Guide for Field Identification. 2012. Available online: http://hdl.handle.net/10883/1115 (accessed on 21 February 2022).
- Orhun, G.E. Maize for life. Int. J. Food Sci. Nutr. Eng. 2013, 3, 13–16.
- Subedi, S. A review on important maize diseases and their management in Nepal. J. Maize Res. Dev. 2015, 1, 28–52.
- Van Nguyen, N.; Ferrero, A. Meeting the Challenges of Global Rice Production. Paddy Water Environ. 2006, 4, 1–9.
- Yoshida, S. Fundamentals of Rice Crop Science; International Rice Research Institute: Los Baños, Philippines, 1981.
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14.
- Phadikar, S.; Sil, J.; Das, A.K. Rice diseases classification using feature selection and rule generation techniques. Comput. Electron. Agric. 2013, 90, 76–85.
- Bekele, B.; Alemayehu, F.; Lakew, B. Food barley in Ethiopia. In Food Barley: Importance, Uses, and Local Knowledge; ICARDA: Aleppo, Syria, 2005; pp. 53–82.
- Mulatu, B.; Grando, S. Barley Research and Development in Ethiopia; EIAR: Addis Ababa, Ethiopia, 2011.
- Paulitz, T.C.; Steffenson, B.J. Biotic stress in barley: Disease problems and solutions. In Barley Production, Improvement, and Uses; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp. 307–354.
- Ramatoulaye, F.; Mady, C.; Fallou, S. Production and use sorghum: A literature review. J. Nutr. Health Food Sci. 2016, 4, 1–4.
- Rooney, W.L. Sorghum. In Cellulosic Energy Cropping Systems; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 109–129.
- Pande, S.; Harikrishnan, R.; Alegbejo, M.; Mughogho, L.; Karunakar, R.; Ajayi, O. Prevalence of sorghum diseases in Nigeria. Int. J. Pest Manag. 1993, 39, 297–303.
- Bao, W.; Zhao, J.; Hu, G.; Zhang, D.; Huang, L.; Liang, D. Identification of wheat leaf diseases and their severity based on elliptical-maximum margin criterion metric learning. Sustain. Comput. Inform. Syst. 2021, 30, 100526.
- Guo, A.; Huang, W.; Ye, H.; Dong, Y.; Ma, H.; Ren, Y.; Ruan, C. Identification of wheat yellow rust using spectral and texture features of hyperspectral images. Remote Sens. 2020, 12, 1419.
- Huang, L.; Ding, W.; Liu, W.; Zhao, J.; Huang, W.; Xu, C.; Zhang, D.; Liang, D. Identification of wheat powdery mildew using in-situ hyperspectral data and linear regression and support vector machines. J. Plant Pathol. 2019, 101, 1035–1045.
- Huang, L.; Wu, Z.; Huang, W.; Ma, H.; Zhao, J. Identification of fusarium head blight in winter wheat ears based on fisher’s linear discriminant analysis and a support vector machine. Appl. Sci. 2019, 9, 3894.
- Khan, I.H.; Liu, H.; Li, W.; Cao, A.; Wang, X.; Liu, H.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; et al. Early Detection of Powdery Mildew Disease and Accurate Quantification of Its Severity Using Hyperspectral Images in Wheat. Remote Sens. 2021, 13, 3612.
- Bohnenkamp, D.; Behmann, J.; Mahlein, A.K. In-field detection of yellow rust in wheat on the ground canopy and UAV scale. Remote Sens. 2019, 11, 2495.
- Xiao, Y.; Dong, Y.; Huang, W.; Liu, L.; Ma, H. Wheat Fusarium Head Blight Detection Using UAV-Based Spectral and Texture Features in Optimal Window Size. Remote Sens. 2021, 13, 2437.
- Sood, S.; Singh, H. An implementation and analysis of deep learning models for the detection of wheat rust disease. In Proceedings of the 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS), Thoothukudi, India, 3–5 December 2020; pp. 341–347.
- Mukhtar, H.; Khan, M.Z.; Khan, M.U.G.; Younis, H. Wheat Disease Recognition through One-shot Learning using Fields Images. In Proceedings of the 2021 International Conference on Artificial Intelligence (ICAI), Islamabad, Pakistan, 5–7 April 2021; pp. 229–233.
- Kumar, D.; Kukreja, V. N-CNN Based Transfer Learning Method for Classification of Powdery Mildew Wheat Disease. In Proceedings of the 2021 International Conference on Emerging Smart Computing and Informatics (ESCI), Pune, India, 5–7 March 2021; pp. 707–710.
- Aboneh, T.; Rorissa, A.; Srinivasagan, R.; Gemechu, A. Computer Vision Framework for Wheat Disease Identification and Classification Using Jetson GPU Infrastructure. Technologies 2021, 9, 47.
- Hussain, A.; Ahmad, M.; Mughal, I.A. Automatic Disease Detection in Wheat Crop using Convolution Neural Network. In Proceedings of the 4th International Conference on Next, Generation Computing, Dehradun, India, 21–22 November 2018.
- Lu, J.; Hu, J.; Zhao, G.; Mei, F.; Zhang, C. An in-field automatic wheat disease diagnosis system. Comput. Electron. Agric. 2017, 142, 369–379.
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128.
- Chen, J.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Identification of rice plant diseases using lightweight attention networks. Expert Syst. Appl. 2021, 169, 114514.
- Wang, Y.; Wang, H.; Peng, Z. Rice diseases detection and classification using attention based neural network and bayesian optimization. Expert Syst. Appl. 2021, 178, 114770.
- Liang, W.j.; Zhang, H.; Zhang, G.f.; Cao, H.x. Rice blast disease recognition using a deep convolutional neural network. Sci. Rep. 2019, 9, 1–10.
- Rahman, M.A.; Shoumik, M.S.N.; Rahman, M.M.; Hena, M.H. Rice Disease Detection Based on Image Processing Technique. In Smart Trends in Computing and Communications: Proceedings of SmartCom 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 135–145.
- Ramesh, S.; Vydeki, D. Rice Disease Detection and Classification Using Deep Neural Network Algorithm. In Micro-Electronics and Telecommunication Engineering; Springer: Singapore, 2020; pp. 555–566.
- Saha, S.; Ahsan, S.M.M. Rice Disease Detection using Intensity Moments and Random Forest. In Proceedings of the 2021 International Conference on Information and Communication Technology for Sustainable Development (ICICT4SD), Dhaka, Bangladesh, 27–28 February 2021; pp. 166–170.
- Chen, J.; Zhang, D.; Nanehkaran, Y.A.; Li, D. Detection of rice plant diseases based on deep transfer learning. J. Sci. Food Agric. 2020, 100, 3246–3256.
- Kamrul, M.H.; Paul, P.; Rahman, M. Machine vision based rice disease recognition by deep learning. In Proceedings of the 2019 22nd International Conference on Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 18–20 December 2019; pp. 1–6.
- Hasan, M.J.; Mahbub, S.; Alom, M.S.; Nasim, M.A. Rice disease identification and classification by integrating support vector machine with deep convolutional neural network. In Proceedings of the 2019 1st International Conference on Advances in Science, Engineering and Robotics Technology (ICASERT), Dhaka, Bangladesh, 3–5 May 2019; pp. 1–6.
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Deep feature based rice leaf disease identification using support vector machine. Comput. Electron. Agric. 2020, 175, 105527.
- Zhou, G.; Zhang, W.; Chen, A.; He, M.; Ma, X. Rapid detection of rice disease based on FCM-KM and faster R-CNN fusion. IEEE Access 2019, 7, 143190–143206.
- Agarwal, R.; Sharma, H. Enhanced Convolutional Neural Network (ECNN) for Maize Leaf Diseases Identification. In Smart Innovations in Communication and Computational Sciences; Springer: Berlin/Heidelberg, Germany, 2021; pp. 297–307.
- Sibiya, M.; Sumbwanyambe, M. A computational procedure for the recognition and classification of maize leaf diseases out of healthy leaves using convolutional neural networks. AgriEngineering 2019, 1, 119–131.
- Barman, U.; Sahu, D.; Barman, G.G. A Deep Learning Based Android Application to Detect the Leaf Diseases of Maize. In Proceedings of the Sixth International Conference on Mathematics and Computing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 275–286.
- Hasan, M.J.; Alom, M.S.; Dina, U.F.; Moon, M.H. Maize Diseases Image Identification and Classification by Combining CNN with Bi-Directional Long Short-Term Memory Model. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP), Dhaka, Bangladesh, 5–7 June 2020; pp. 1804–1807.
- Xu, Y.; Zhao, B.; Zhai, Y.; Chen, Q.; Zhou, Y. Maize Diseases Identification Method Based on Multi-Scale Convolutional Global Pooling Neural Network. IEEE Access 2021, 9, 27959–27970.
- Tian, J.; Zhang, Y.; Wang, Y.; Wang, C.; Zhang, S.; Ren, T. A method of corn disease identification based on convolutional neural network. In Proceedings of the 2019 12th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 14–15 December 2019; Volume 1, pp. 245–248.
