Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Climate change and the need to feed an increasing population undermines food production and safety, representing the reasons behind the development of a new agriculture that is much more sustainable, productive and accessible worldwide. Genome editing and, in particular, clustered regularly interspaced palindromic repeats/CRISPR-associated protein (CRISPR/Cas) tools will play a major role in plant breeding to address these concerns. CRISPR/Cas includes a series of genome editing tools relying on the recognition and cleavage of target DNA/RNA sequences to introduce specific mutations.
- CRISPR
- crop improvement
- genetic variability
- stress tolerance
- food quality
- synthetic biology
- sustainable agriculture
1. Introduction
The term “sustainable agriculture” encloses practices of farming addressed to the production of high-quality and safe agricultural products without compromising natural environments and the social and economic conditions of farmers. According to the Agricultural Sustainability Institute at UC Davis (https://www.nal.usda.gov/legacy/afsic/sustainable-agriculture-definitions-and-terms, accessed on 10 January 2022), the main goal of sustainable agriculture remains the preservation of the ability of future generations to meet their own needs, ensuring inclusive economic growth.
In the era of climate change, environmental threats will affect farmers at both the economic and the agronomic level, influencing crop yield and quality and with further negative effects on plant resistance to both abiotic and biotic stress. In order to counteract these environmental threats, and to have the chance to reach a sustainable production of industrial manufacture, new technologies will be used on the basis of the current knowledge in the biotechnology field.
Clustered regularly interspaced palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) represent a new perspective for genetic engineering and the last frontiers of the new breeding techniques (NBTs) and genome editing (GE) tools.
1.1. CRISPR/Cas System
CRISPR/Cas systems are part of the adaptative immune system of archaea and bacteria in ensuring protection against viruses. The mechanism of action relies on the recognition and the cleavage of foreign DNA or RNA of invading viruses. One of the main characteristics is the integration of short fragments of the invading DNA (spacers) into the CRISPR locus, conferring the heritable immunity of bacteria. The CRISPR locus consists of an array of unique spacer sequences, derived from foreign invading DNA and interspaced by identical repeat sequences (crisprRNA or crRNA), along with a sequence encoding for the trans-activating crRNA (tracrRNA) and a series of genes encoding CRISPR-associated (Cas) endonucleases, responsible for the cleavage of the genetic material. The transcription of the CRISPR locus allows for the formation of a single mRNA (pre-crRNA), which is partially complementary to tracrRNA, leading to the formation of an RNA duplex. The RNase III recognizes the RNA duplex and cleaves the double-stranded RNA (dsRNA) to form crRNA–tracrRNA complexes that activate and drive the Cas protein to the target sequence. The Cas protein is capable of introducing a double-strand break (DSB) only in the presence of a short conserved protospacer-adjacent motif (PAM) downstream of the target DNA, representing an essential prerequisite for the recognition of the target sequence [1][2].
CRISPR/Cas systems have been divided into two classes, six types and several subgroups based on Cas proteins and the nature of the interference complex [3]. To date, the most common type, used as a genome editing tool, is the Type II Cas9 from Streptococcus pyogenes (SpCas9). The Cas9 protein consists of a bi-lobed architecture, including a large recognition (REC) lobe and a small nuclease (NUC) lobe. The NUC lobe includes a protospacer-adjacent motif (PAM)-interacting domain (PI) and two cleavage domains known as RuvC and HNH domains, each of which cleaves one strand of the target DNA three nucleotides upstream of the PAM sequence (Figure 1A) [4][5]. The REC lobe contributes toward activating Cas proteins when combined with the tracrRNA–crRNA complex.
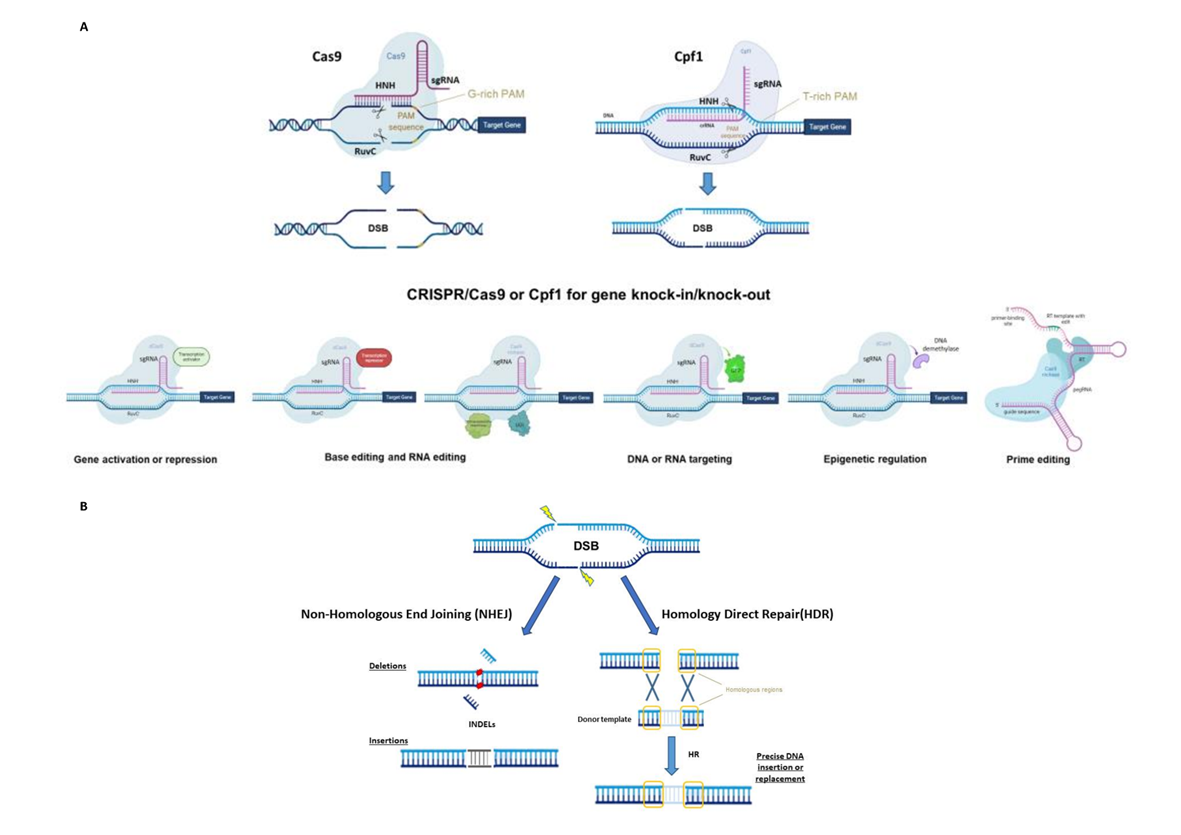
Figure 1. (A) Principal CRISPR/Cas9 or Cpf1 types of editing (top) and possible use for gene knock-in or knock-out (bottom), synthetic-guide RNA (sgRNA), double-strand break (DSB), protospacer-adjacent motif (PAM), reverse transcriptase (RT), uracil DNA glycosylase inhibitor (UGI), green fluorescent protein (GFP) and prime editing gRNA (pegRNA). (B) The two main DNA repair mechanisms and genetic mutations.
The use of the CRISPR/Cas system as a genomic engineering tool occurred when Jinek et al., 2012 [4] (Figure 2) showed that the target DNA sequence could be reprogrammed simply using a chimeric synthetic-guide RNA (sgRNA), obtained by the fusion of crRNA and tracrRNA sequences and changing the 20 nucleotides of the crRNA that confer the targeting specificity. Once the Cas9 combined with the sgRNA recognizes the complementary spacer sequence adjacent to PAMs on double-strand DNA, genetic modifications are produced through the induction of DSB (Figure 1A), followed by the activation of DNA repair mechanisms (Figure 1B) through non-homologous end-joining (NHEJ) or homologous direct repair (HDR) [6][7][8].
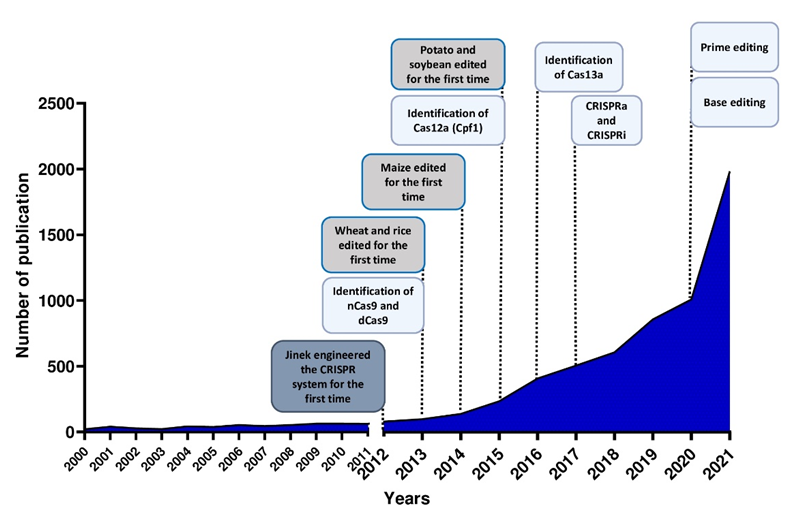
Figure 2. Number of publications in plant breeding using CRISPR systems and major achievements in the last 10 years.
1.2. CRISPR Variants, Orthologs and Engineered Systems
New types of Class II CRISPR/Cas systems are continuously found in bacteria, showing different features, and customized for use in plants (Figure 2). Several Cas9 orthologs, with different PAM specificities from other bacteria, have been discovered and some of them have already been applied for genome editing in model species [9]. Furthermore, it has been demonstrated that mutations in the PAM-interacting domains of wild-type SpCas9 could lead to SpCas9-engineered variants recognizing different PAM sequences [10], thus increasing the spectrum of editable target sites in the genome.
For example, Cas13a (C2c2) enzyme belongs to the Type VI Class II (Figure 1A). It is specialized in RNA recognition and cleavage, enabling its use in post-transcriptional repression, degradation of genetic material of RNA viruses and RNA binding [11]. Two studies have demonstrated the activity of Cas13a in rice protoplast to knock down endogenous genes and in Nicotiana benthamiana plants for interfering against RNA viruses [12][13]. Otherwise, the Cas12a (Cpf1) enzyme belongs to Class II Type V (Figure 1A) and differs from Cas9 in several important features: it recognizes the target region through PAM favoring AT-rich regions (5′-TTTN-3′), it cleaves the target sequence by producing DNA ends with a 5′ overhang and the crRNA directly functions as a guide RNA without the need for a complex with tracrRNA to be processed. This Cas protein has been used in many crop species [14] since its first application in rice and tobacco [15].
Furthermore, the Cas9 protein has been re-engineered through point mutations in RuvC and/or HNH nuclease domains, inactivating the catalytic activity of each domain (Figure 1A). It can be exploited to produce a nickase (nCas9) or a dead enzyme (dCas9) with complete loss of DNA cleavage activity. The nCas9 is usually used to enhance the specificity of CRISPR/Cas9, combining two nCas9 with pairs of sgRNAs that respectively cut only one DNA strand and therefore increase the number of recognized target bases. The dCas9 protein can operate as cargo to load and deliver proteins with different functions to a specific target site. The use of inactive enzymes (nCas9 or dCas9) can also facilitate the directional introduction of DNA fragments at a specific site and the assembly of base and prime editors (discussed below). This enzyme has been employed in base editing as well as in genetic and epigenetic regulation of gene expression [16]. Otherwise, the most frequent use of dCas9 is in the activation (CRISPRa, activator) and repression (CRISPRi, interfering) of gene expression without introducing mutations in the genome (Figure 1A and Figure 2). Following this approach, the dCas9, guided by a sgRNA to a specific regulatory region, is fused to transcriptional modulators, which are generally transcriptional factors or protein domains recruiting key regulatory elements to control gene expression. Their use has been reported in plants both as activators [17][18][19][20] and as repressors [21][22]. The dCas9, associated with an acetyltransferase or a methyltransferase, can also act at the epigenetic level as CRISPRi and CRISPRa systems [23][24].
Both nCas9 and dCas9 are involved in the assembly of base editing and prime editing systems. Base editors, including cytosine base editors (CBEs) and adenosine base editors (ABEs), catalyze C/G to T/A or A/T to G/C transitions in DNA or RNA molecules (Figure 1A and Figure 2). They have been optimized for plant genomes, providing high efficiency and precise editing at single base resolution [25]. Base editing systems have been applied in major crops such as wheat, rice, maize, tomato, potato, soybean and rapeseed [26][27]. Prime editing enables rewriting of genetic information into a specified DNA target site using a reverse transcriptase (RT) fused to a nickase enzyme and a prime editing guide RNA (pegRNA) to copy genetic information directly into the target genomic sequence (Figure 1A and Figure 2) [28]. Although prime editing still needs to be improved for editing efficiency in plants, it has been applied to obtain precise modifications in rice, maize, potato and tomato [27][28][29], especially using the second generation of prime editors (PE2), in which an engineered RT with improved features (such as increased processivity, substrate affinity and inactivated RNase H activity) is fused to an nCas9 [28][30].
1.3. New Perspectives for the Use of CRISPR/Cas System
CRISPR opens many doors for plant breeders to boost breeding programs towards ambitious targets, thanks not only to the feasibility of its application in a wide range of crop species but also to its versatility as a genetic tool that is constantly evolving. The use of CRISPR-engineered systems can foster the generation of a wide range of heritable genetic mutations such as In/Dels, targeted insertions, point mutations and nucleotide substitutions that are the most frequent modifications obtained, as well as targeted chromosomal rearrangements and genetic or epigenetic control of gene expression (Figure 1A). In addition, it offers the advantage of decreasing off-targets and pleiotropic effects, without neglecting the possibility of obtaining transgene-free edited plants.
The possibility of producing transgene-free plants has been exploited through self-pollination and segregation of exogenous DNA [31], transiently expressing a plasmid vector encoding for Cas9 and gRNAs [32][33] or through the delivery of pre-assembled CRISPR/Cas9 ribonucleoproteins (RNPs) [34]. By delivering the Cas9 protein instead of the vector, there is no transfer of specific DNA from one species to another. In addition, the Cas9 protein remains inside the cells for three/five days and then is degraded, also reducing the off-target events. All together, these approaches will make CRISPR a useful tool for a new generation of plants that potentially do not fall within the scope of the current regulation process of genetically modified (GM) products.
Moreover, the discovery and the improvement of CRISPR as a precise genome editing tool has resulted in the establishment of several CRISPR-based companies that are hoping to capitalize on this new technology. Indeed, the potential of gene editing to address the 21st century’s problems has taken hold in the agricultural industry and the list of CRISPR companies is growing each day. As an example, Synthetic Genomics® (https://www.viridos.com/, accessed on 17 November 2021) uses synthetic biology solutions to produce microalgae with higher levels of lipids to be used to address global sustainability problems. Plantedit® (https://plantedit.com/, accessed on 17 November 2021) uses genome editing to generate modified soybean with a high oil content. Pairwise Plants (https://www.pairwise.com/, accessed on 17 November 2021) is currently developing edited plants to assist farmers by providing them with new varieties of crops that require fewer resources to grow. Inari Agriculture® (https://inari.com/, accessed on 17 November 2021) is using CRISPR to enhance plant breeding, by managing specific gene expression in plants, to develop customized seeds. Hudson River Biotechnology® (https://www.hudsonriverbiotechnology.com/, accessed on 17 November 2021) employs CRISPR technology to edit plants and microorganisms through a validated molecular breeding workflow called TiGER (Target identification, Guide selection, Entry into the cell and Regeneration). Yield10® Bioscience (https://www.yield10bio.com/, accessed on 17 November 2021) aims to improve the yield of crops such as canola and soybeans, and also to increase the oil content of these and other oilseeds. Other relevant companies are Benson Hill Biosystems® (https://bensonhill.com/, accessed on 17 November 2021), Corteva® (https://www.corteva.com/, accessed on 17 November 2021) (agricultural division of DowDuPont), Syngenta® (https://www.syngentagroup.com/, accessed on 17 November 2021) and Tropic Biosciences® (https://www.tropicbioscience.com/, accessed on 17 November 2021).
Here, researchers focus on the recent advances in CRISPR technology for the improvement of the most cultivated crop species and its potential applications in synthetic biology (Figure 2), with particular regard to traits such as quality, yield and stress tolerance. Lastly, researchers report a quick focus on the global regulatory framework on GM plant legislation.
This entry is originated from: 10.3390/encyclopedia2010036
References
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 80, 346.
- Fichtner, F.; Urrea Castellanos, R.; Ülker, B. Precision Genetic Modifications: A New Era in Molecular Biology and Crop Improvement. Planta 2014, 239, 921–939.
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR-Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–822.
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52.
- Wyman, C.; Kanaar, R. DNA Double-Strand Break Repair: All’s Well That Ends Well. Annu. Rev. Genet. 2006, 40, 363–383.
- Podevin, N.; Davies, H.V.; Hartung, F.; Nogué, F.; Casacuberta, J.M. Site-Directed Nucleases: A Paradigm Shift in Predictable, Knowledge-Based Plant Breeding. Trends Biotechnol. 2013, 31, 375–383.
- Chen, K.; Gao, C. Targeted Genome Modification Technologies and Their Applications in Crop Improvements. Plant Cell Rep. 2014, 33, 575–583.
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259.
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 System for Plant Genome Editing: Current Approaches and Emerging Developments. Agronomy 2020, 10, 1033.
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two Distinct RNase Activities of CRISPR-C2c2 Enable Guide-RNA Processing and RNA Detection. Nature 2016, 538, 270–273.
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA Targeting with CRISPR-Cas13. Nature 2017, 550, 280–284.
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA Virus Interference via CRISPR/Cas13a System in Plants. Genome Biol. 2018, 19, 1–9.
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A Versatile Tool in the Plant Genome Editing Tool Box for Agricultural Advancement. Front. Plant Sci. 2020, 11, 1–17.
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient Targeted Mutagenesis of Rice and Tobacco Genomes Using Cpf1 from Francisella Novicida. Sci. Rep. 2016, 6, 1–9.
- Moradpour, M.; Abdulah, S.N.A. CRISPR/DCas9 Platforms in Plants: Strategies and Applications beyond Genome Editing. Plant Biotechnol. J. 2020, 18, 32–44.
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.F. A Potent Cas9-Derived Gene Activator for Plant and Mammalian Cells. Nat. Plants 2017, 3, 930–936.
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and MTALE-Act Systems. Mol. Plant 2018, 11, 245–256.
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for Highly Efficient Multiplexed Gene Activation in Plants. Nat. Plants 2021, 7, 942–953.
- Malzahn, A.; Zhang, Y.; Qi, Y. CRISPR-Act2.0: An Improved Multiplexed System for Plant Transcriptional Activation. In Plant Genome Editing with CRISPR Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 83–93.
- Piatek, A.A.; Lenaghan, S.C.; Neal Stewart, C. Advanced Editing of the Nuclear and Plastid Genomes in Plants. Plant Sci. 2018, 273, 42–49.
- Vazquez-Vilar, M.; Bernabé-Orts, J.M.; Fernandez-del-Carmen, A.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. A Modular Toolbox for GRNA-Cas9 Genome Engineering in Plants Based on the GoldenBraid Standard. Plant Methods BioMed Cent. 2016, 12, 1–12.
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-Based Tools for Targeted Transcriptional and Epigenetic Regulation in Plants. PLoS ONE 2019, 14, 1–17.
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; de de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved Drought Stress Tolerance in Arabidopsis by CRISPR/DCas9 Fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9.
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in Agriculture and Plant Biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677.
- Ren, Q.; Sretenovic, S.; Liu, G.; Zhong, Z.; Wang, J.; Huang, L.; Tang, X.; Guo, Y.; Liu, L.; Wu, Y.; et al. Improved Plant Cytosine Base Editors with High Editing Activity, Purity, and Specificity. Plant Biotechnol. J. 2021, 19, 2052–2068.
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise Plant Genome Editing Using Base Editors and Prime Editors. Nat. Plants 2021, 7, 1166–1187.
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157.
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime Genome Editing in Rice and Wheat. Nat. Biotechnol. 2020, 38, 582–585.
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043.
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci. Rep. 2017, 7, 1–6.
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.W.; et al. Marker-Free Carotenoid-Enriched Rice Generated through Targeted Gene Insertion Using CRISPR-Cas9. Nat. Commun. 2020, 11, 1–10.
- Waltz, E. Gene-Edited CRISPR Mushroom Escapes US Regulation. Nature 2016, 532, 293.
- Globus, R.; Qimron, U. A Technological and Regulatory Outlook on CRISPR Crop Editing. J. Cell. Biochem. 2018, 119, 1291–1298.
This entry is offline, you can click here to edit this entry!
