Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Area Studies
CP4-EPSPS (Agrobacterium sp. strain CP4 5-enolpyruvylshikimate-3-phosphate synthase) protein showed remarkable thermostability and was highly resistant to proteases, such as trypsin. However, some proteases are able to degrade CP4-EPSPS efficiently. Therefore, these proteases may play key roles in elimintating the pollution of CP4-EPSPS to the environment.
- CP4-EPSPS
- protease
- Stenotrophomonas maltophilia
1. Introduction
GM (Genetically Modified) crops have been planted worldwide over the past decades. During 1996 to 2016, the acreage of GE (Genetically Engineered) crops expanded 109-fold. Among them, the crop covering the most hectares globally is soybean. In 2016, 78% of soybeans planted were GE varieties [1]. There are 30 events of transgenic soybeans, and one of the most representative transgenic soybeans is the Roundup Ready soybean (RRS, event GTS40-3-2). The transgenically expressed protein in RRS is CP4-EPSPS protein, a variant of plant 5-enolpyruvyl-shikimate-3-phosphate synthase (EPSPS). Plant EPSPS is involved in the Shikimate pathway, which plays a critical role in many plant functions, such as the production of aromatic amino acids [2,3]. CP4-EPSPS was originally isolated from the soil Agrobacterium sp. strain CP4. The full length of this gene encodes a protein of 55.6 kDa, including a 72 aa chloroplast transit peptide and a mature protein of 47.6 kDa. As a variant of plant EPSPS, it has a lower binding affinity to glyphosate [4]. Therefore, constitutive expression of CP4-EPSPS in crops confers increased tolerance to the Roundup family of agricultural herbicides. Except for soybean [5,6], CP4-EPSPS was also applied in transgenic rice [7], cotton (event MON 1445, MON 88913) [8], corn, etc.
Even though the commercial use of GM crops has increased dramatically, the safety of GM plants is still a controversial topic, attracting both the attention of the scientific community and the public. Therefore, the effects of transgenic plants on domestic animals and the environment have been intensively studied [9]. CP4-EPSPS shows no obvious similarity with proteins associated with allergy and toxicity [10], and several investigations indicated that doses of CP4-EPSPS much higher than the dietary supplement level had no obvious toxicological effect on the reproductive function in rats [11,12] or animal health as a dietary supplementation [13]. However, some research has still raised possibilities that these plants may cause unintended effects via different pathways, which have to be systematically evaluated. Therefore, any genetically modified plants or animals should undergo strict safety evaluation before entering the food market. Previous studies indicated that CP4-EPSPS protein is very stable, and a trace amount of the transgenic protein was still detectable in the highly processed RRS soybean products [14]. Moreover, a higher level of CP4-EPSPS was observed in leaf tissues than in the seeds and roots [10].
2. Recombinant Expression of CP4-EPSPS
E. coli BL21 (DE3) bearing pET28a-EPSPS, pET28a-His6-EPSPS, and pET28a-EPSPS-His6 were used to express the recombinant proteins. The ORF of CP4-EPSPS is 1623 bp (Genbank accession No. NP_740524) and encodes a protein of approximately 47 kDa. The result of SDS–PAGE indicated that a band consistent with the predicted size of CP4-EPSPS was detected in the supernatant of the cell lysate (Figure 1a). The target proteins bearing 6 × His tags were purified with Ni-NTA. Meanwhile, the recombinant CP4-EPSPS without a fusion tag was purified with native gel. The result of SDS–PAGE indicated that all of them were purified successfully (Figure 1b–d).
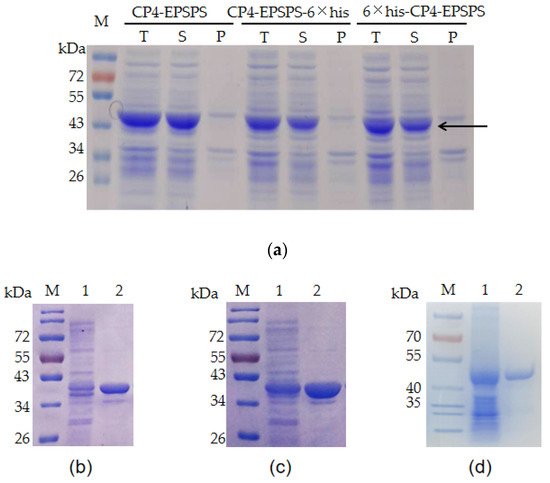
Figure 1. Expression and purification of recombinant CP4-EPSPS protein. (a) Expression of recombinant CP4-EPSPS with E. coli BL21 (DE3) as a host. T: total protein of the cells; S: supernatant of the cell lysate; P: pellet of the cell lysate. The target protein is indicated with an arrow. (b) Purified 6 × his-CP4-EPSPS; (c) purified CP4-EPSPS-6 × his; (d) purified CP4-EPSPS. Lane 1: supernatant of the cell lysate; Lane 2: purified target protein; M: protein molecular weight marker (the size of each band is indicated on the left).
3. Stability of the Recombinant CP4-EPSPS
To investigate the stability of CP4-EPSPS, the recombinant protein was diluted to 2 mg/mL and incubated at room temperature. The result of SDS–PAGE indicated that no obvious degradation of the target protein was detected in 25 days (Figure 2a). The three forms of recombinant proteins were also boiled in a water bath and no obvious degradation of the target protein was detected after 1.5 h of heating (Figure 2b). In addition, the target protein remained soluble after 30 min of microwaving (Figure 2c). These results indicated that CP4-EPSPS was highly resistant to heat. To investigate the effect of protease on CP4-EPSPS, commercial trypsin was used to digest CP4-EPSPS. The result indicated that CP4-EPSPS was resistant to high concentrations of trypsin. It took 96 h for trypsin to degrade CP4-EPSPS completely at a molar ratio of 1:10 (Figure 3).
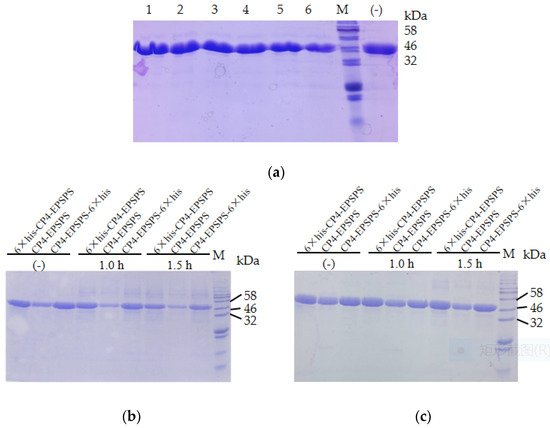
Figure 2. Stability of the recombinant CP4-EPSPS. (a) Stability of the recombinant CP4-EPSPS at room temperature. Lane 1–6: recombinant 6 × his-CP4-EPSPS incubated at room temperature for 1, 2, 4, 7, 15, and 25 days, respectively; (−) the negative control. (b) Stability of the recombinant CP4-EPSPS with boiling. (c) Stability of the recombinant CP4-EPSPS with microwaving; M, protein molecular weight marker (the size of each band is indicated on the right).
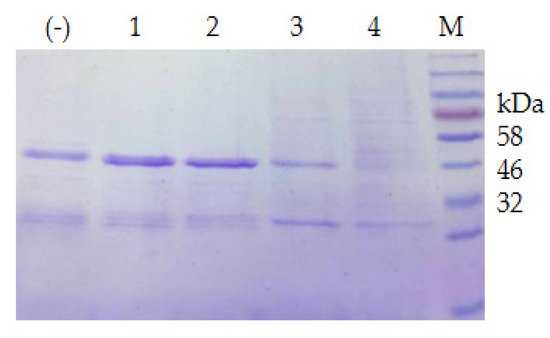
Figure 3. Digestion of CP4-EPSPS with trypsin. Lane 1–4: digestion of 6 × his-CP4-EPSPS with trypsin at a molar ratio of 10:1 for 12, 24, 84, and 96 h, respectively; (−) the negative control; M, protein molecular weight marker (the size of each band is indicated on the right).
4. Screening Psychrophilic Bacteria Strains Capable of Degrading the Recombinant CP4-EPSPS
Twenty-one psychrophilic bacterial strains isolated from the south polar region were inoculated on ½R2A plates supplemented with 1% milk and incubated at 18 °C. Clear halos appeared around the colonies of CCTCC2016780, 741, 742, 743, and 808, while a relatively small halo formed around CCTCC2016809 after 3 days (Figure 4), which indicated that these strains secreted cold-active proteases to the surrounding environment. The experiment was run in triplicate. CCTCC2016780, 741, 742, 743, and 808 exhibited robust extracellular protease activities and clear halos were formed in all three repeats. These strains were incubated in 1/2R2A supplemented with 1% milk for 3 days. The supernatants of the cell cultures were gathered and incubated with the recombinant CP4-EPSPS. After 48 h of digestion, proteases secreted by CCTCC2016780 and 808 showed obvious degradation of CP4-EPSPS and the band of CP4-EPSPS almost disappeared. The extracellular proteases of CCTCC2016741 and 742 were also capable of degrading CP4-EPSPS and the target protein was partially digested. On the contrary, CCTCC2016809 exhibited no obvious effect on CP4-EPSPS (Figure 5). Since CCTCC2016780 demonstrated the most obvious effect, it was used for further study.
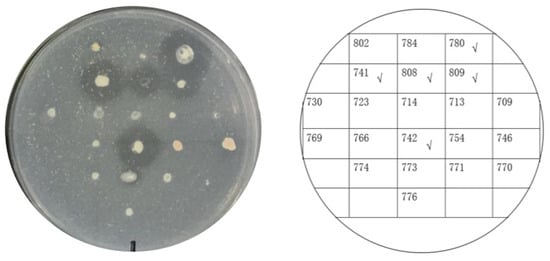
Figure 4. Screening psychrophilic bacterial strains with extracellular protease activity. The serial number of each strain is indicated on the right. Strains with obvious extracellular protease activity are ticked.
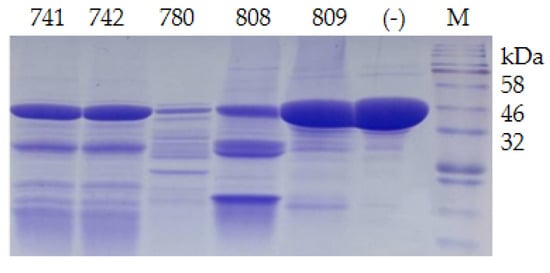
Figure 5. Degradation of recombinant CP4-EPSPS with extracellular proteases of psychrophilic bacterial strains. The serial number of each strain is indicated on the SDS–PAGE gel; (−) the negative control; M, protein molecular weight marker (the size of each band is indicated on the right).
5. Identification of Psychrophilic Bacteria CCTCC2016780
According to the molecular taxonomy result of CCTCC, CCTCC2016780 belongs to Mucilaginibacter gotjawali. During the process of obtaining a pure culture, we realized that this isolate was composed of two strains. Both strains formed white colonies on 1/2R2A plates after 48 h of incubation at 18 °C (Figure 6). Both strains were identified by 16S rDNA sequence analysis. The strain that showed robust growth was identified as Pseudomonas sp., since its 16S rDNA had 100% identity with Pseudomonas moraviensis WTB8 and Pseudomonas sp. PS13. The 16S rDNA of the petite colonies showed 99% identity with Stenotrophomonas maltophilia 7K14 and R551-3, which suggested that it belonged to Stenotrophomonas maltophilia. Accordingly, these two strains were named Pseudomonas sp. 780 and S. maltophilia 780, respectively.
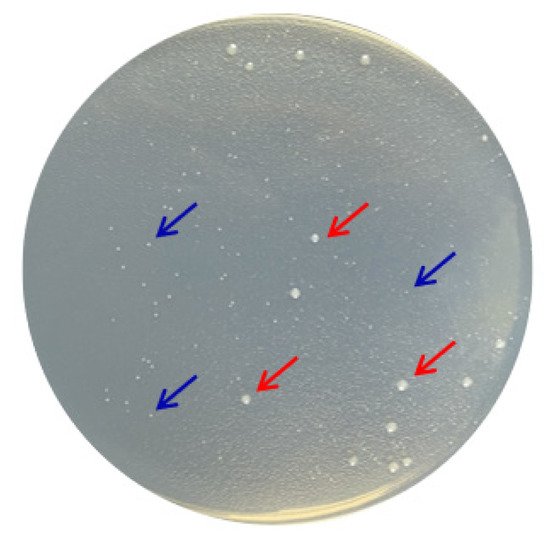
Figure 6. Two bacterial strains isolated from Mucilaginibacter gotjawali 780. The red arrows indicate Pseudomonas sp. 780 with robust growth and the blue arrows indicate S. maltophilia 780, which formed petite colonies.
The extracellular proteases of both strains were induced with milk and utilized for the degradation of the recombinant CP4-EPSPS. The result of SDS–PAGE indicated that the extracellular proteases of S. maltophilia 780 degraded CP4-EPSPS efficiently, and almost all CP4-EPSPS was degraded in 5 h (Figure 7). On the contrary, the extracellular proteases of P. sp. 780 had no effect on the target protein. Therefore, S. maltophilia 780 was used for further study.
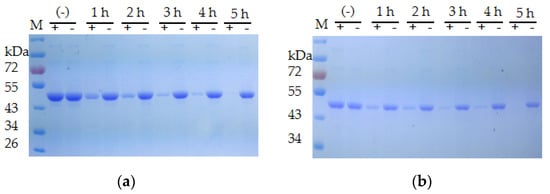
Figure 7. Time course of the degradation of recombinant CP4-EPSPS with the extracellular proteases of S. maltophilia 780. (a) Degradation of 6 × HIS-CP4-EPSPS; (b) degradation of CP4-EPSPS-6×HIS; (−) the negative control (degradation with supernatant of cell culture heated at 100 °C for 30 min); M, protein molecular weight marker (the size of each band is indicated on the left).
The morphological study indicated that S. maltophilia 780 was Gram negative and had a short rod shape (Figure 8), which is consistent with the typical features of S. maltophilia. As a psychrophilic bacterium, S. maltophilia 780 showed obvious growth at 18 °C, while its growth was completely inhibited at 37 °C in both 1/2R2A and LB media.
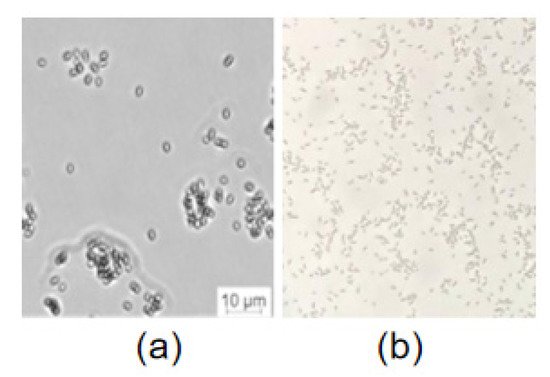
Figure 8. Microscopic examination of S. maltophilia 780. (a) DIC (digital image correlation) with Leica microsystems CMS GmbH; (b) Gram staining.
6. Characteristics of the Extracellular Proteases of S. maltophilia 780
The effect of various nitrogen sources on the biosynthesis of extracellular proteases of S. maltophilia 780 was investigated. The organic nitrogen sources had stronger inducing effects than simple nitrogen sources, such as (NH4)2SO4. Among the organic nitrogen sources, milk demonstrated the most obvious effect (Figure 9). Because S. maltophilia 780 is a psychrophilic bacteria, its extracellular proteases had weak activity at 0 °C and the activity increased with the elevation of the temperature (Figure 10a). It is puzzling that the activity reached the maximum level at 65 °C, which was relatively high in comparison with other psychrophiles. Consistent with the features of cold-active enzymes, the extracellular proteases of S. maltophilia 780 were sensitive to heat and most of the activity was lost at 70 °C in 10 min. On the other hand, they were relatively stable at 25 °C and retained approximately 80% of the activity after 30 min (Figure 10b). The kinetic parameters of the extracellular proteases of S. maltophilia 780 were investigated. The binding affinity (KM) was 767.08 µm. On the contrary, the catalytic turnover (kcat) was 1.39 × 103 s−1 (Figure 10c).
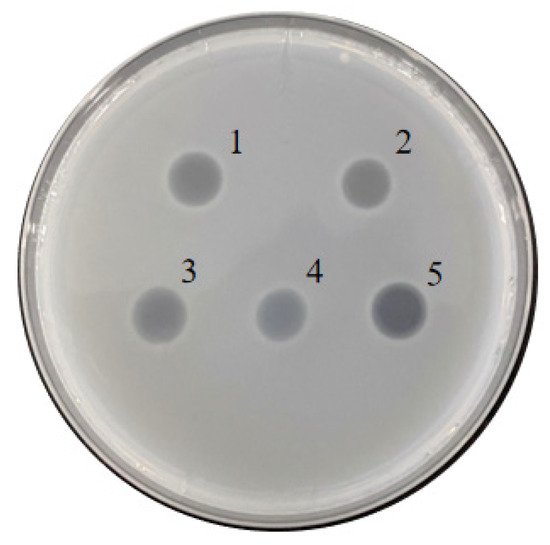
Figure 9. Extracellular protease generated by S. maltophilia 780 induced with different nitrogen sources. 1: extract of Roundup Ready soybean powder; 2: 1% yeast extract; 3: 1% trypton; 4: 1% (NH4)2SO4; 5: 1% milk powder.
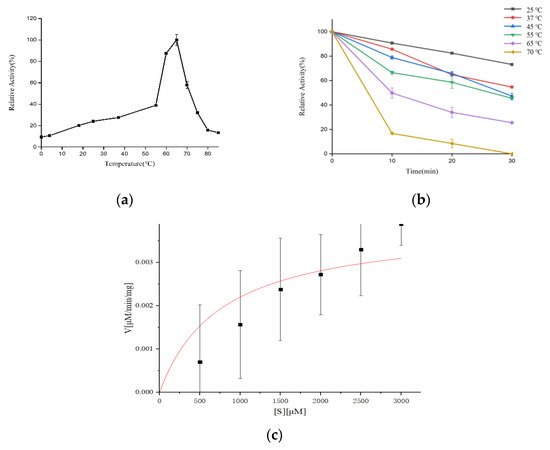
Figure 10. Effect of temperature on the activity and stability of the extracellular proteases of S. maltophilia 780. (a) Optimum temperature of the enzymes; (b) effect of temperature on the thermostability of the enzymes; (c) kinetic analysis of the extracellular proteases of S. maltophilia 780.
7. Degradation of CP4-EPSPS in RRS with S. maltophilia 780a
S. maltophilia 780 was incubated in 1/2R2A supplemented with 1% milk for 3 days, and the supernatant was applied to degrade CP4-EPSPS in RRS extract. The result of the test strips indicated that the concentration of CP4-EPSPS decreased to an undetectable level after 30 h of incubation (Figure 11). As the culture of S. maltophilia 780 could degrade CP4-EPSPS in the transgenic soybean efficiently, this isolate was cultivated in the extract of RRS directly. The result indicated that S. maltophilia 780 was able to grow in the extract of RRS, while E. coli DH5α grew much slower (Figure 12a). Meanwhile, ELISA indicated that CP4-EPSPS in the extract was degraded to an almost undetectable level after approximately 72 h. On the other hand, E. coli DH5α had no obvious effect (Figure 12b).
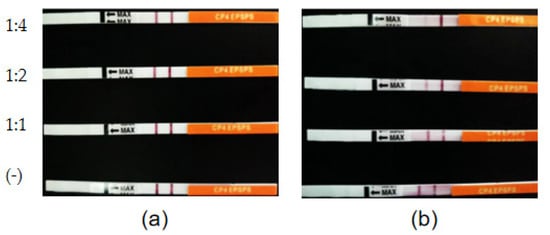
Figure 11. Time course of the degradation of CP4-EPSPS in the extract of RRS using the cell culture of S. maltophilia 780. (a) RRS extract incubated with inactivated enzymes (negative controls). (b) RRS extract incubated with the enzymes. The ratio of the extract to the enzymes (v/v) is indicated on the left of the figure.
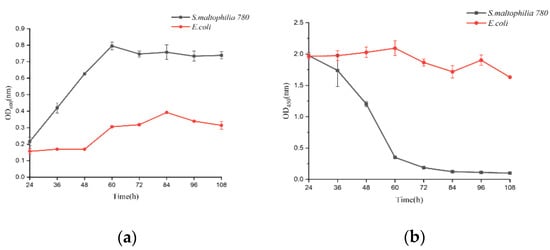
Figure 12. Degradation of CP4-EPSPS in the extract of RRS during the growth of S. maltophilia 780. (a) Growth curve of E. coli and S. maltophilia 780 in the extract of RRS. (b) CP4-EPSPS level in the cell culture.
8. Genomic Sequencing of S. maltophilia 780 and the Genes Encoding Proteases
The general genomic features of S. maltophilia 780 are listed in Table 1. The G + C content of S. maltophilia 780 was 64.58%, which was consistent with that of other S. maltophilia genomes. The genome size of S. maltophilia 780, slightly larger than the previously reported S. maltophilia genomes, was approximately 5.77 Mb with 5536 predicated CDS [16]. Functional annotation indicated that more than a hundred genes coded proteases and peptidases.
Table 1. Genomic features of S. maltophilia 780.
| Feature | Value |
|---|---|
| Genome size/Mb | 5.77 |
| G + C content/% | 64.58% |
| Protein-coding genes (CDS) | 5536 |
| rRNA (5S, 16S, 23S) | 3 |
| tRNA | 55 |
| Other ncRNA | 54 |
This entry is adapted from the peer-reviewed paper 10.3390/biom12020318
This entry is offline, you can click here to edit this entry!
