Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Psychology, Biological
Major depressive disorder (MDD) is the most prevalent, chronic, and complicated neuropsychiatric disorder of the current era, causing 800,000 people to commit suicides annually (WHO, 2020). In MDD, several miRNAs have been identified in disease progression by targeting various pathogenic proteins or associated pathways, and they are dysregulated by pathogenic protein expression.
- cimiRNAs
- depression
1. Introduction
Major depressive disorder (MDD) is the most prevalent, chronic, and complicated neuropsychiatric disorder of the current era [1][2], causing 800,000 people to commit suicides annually (WHO, 2020). It is characterized by negative sequelae such as low mood, lack of interest and pleasure, fluctuations in weight or appetite, sleep disturbances (insomnia or hypersomnia), fatigue, psychomotor retardation or agitation, remorse, impaired concentration, and lower self-confidence, all of which can lead to suicidal thoughts or attempts [3]. Recently, the World Health Organization (WHO) listed depression as the second leading cause of disability (after cancer) that has burdened nations economically and jeopardized life by affecting over 264 million people worldwide, and plausibly predicted that it would be ranked first by 2030 (WHO, 2020) [4]. Hence, this alarming situation has turned MDD into a major public health concern in developed states.
In the past few years, research on circulatory miRNAs (cimiRNAs) as biological indicators of various pathogenic conditions has gained momentum [5]. They travel in bodily fluids such as serum [6], plasma [7], urine [8], cerebrospinal fluid [6], and saliva [9][10] and impart their activity in distant cells or tissues to administer endocrine gene regulation [11]. Notably, several reports have associated dysregulated cimiRNAs with different pathological and psychological conditions, including psychiatric disorders, neurological disorders, cancer, and other metabolic disorders. In comparison to metabolomic [12] and proteomic biomarkers [13], miRNAs propose various advantages over other RNA-based molecules, such as robust stability in bodily fluids [14], specific expression in particular tissues [13], less variation in results between gender, age or time of sampling [15][16], and fast and accurate quantification using routine laboratory methods (RT-qPCR) [17]. Thus, accurate interpretations can be deduced because mature miRNA levels usually align with miRNA activity, whereas post-translational modifications of proteins usually result in complex correlations between protein activity and expression levels. In MDD, several miRNAs have been identified in disease progression by targeting various pathogenic proteins or associated pathways, and they are dysregulated by pathogenic protein expression [18]. Moreover, several reports have presented dysregulation of cimiRNAs in the CSF or blood of MDD patients [19][20].
2. Profiling of Dysregulated cimiRNAs in MDD
Figure 1 show a detailed explanation of the data acquired from the 32 studies that served as the basis for this meta-analysis. These studies included 11 blood, nine plasma, four serum, two CSF + blood, four PBMC, and three exosome reports (Figure 1A). Across all reports, a total of 99 cimiRNAs were found dysregulated in MDD; 20 cimiRNAs (20%) were reported in two or more studies, and 10 cimiRNAs (10%) in three or more studies (Figure 1). Most of these cimiRNAs are highly expressed in the brain tissues and regulate neuronal and MDD-associated processes.
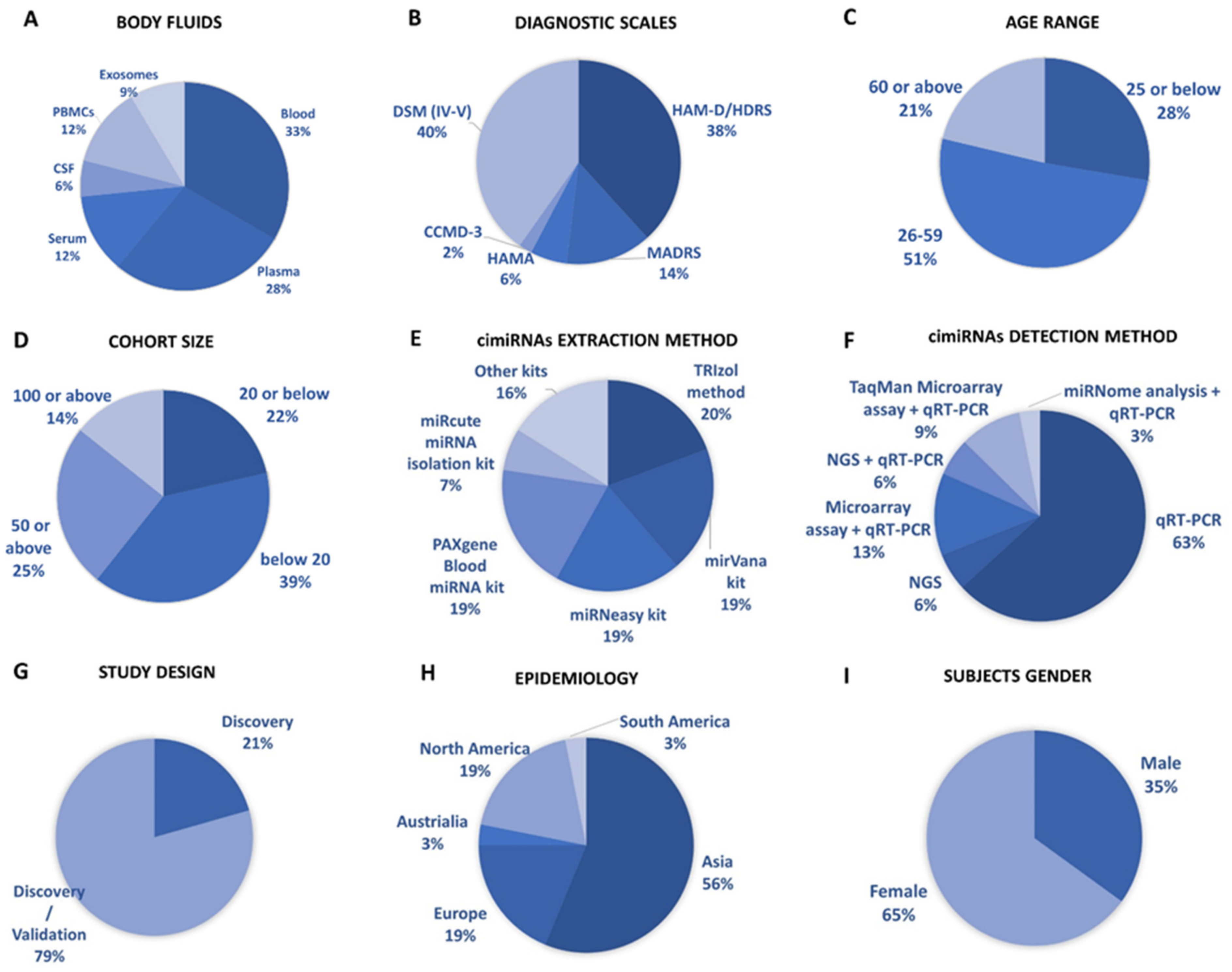
Figure 1. Overview of cimiRNA studies in MDD. (A) Fraction of human body fluids analyzed for cimiRNAs in MDD. (B) Clinical scales used for MDD diagnosis. (C) Age range of participants for MDD studies. (D) Number of MDD samples analyzed. (E) Extraction strategies employed for cimiRNA isolation. (F) Detection strategies used for cimiRNAs. (G) Types of studies illustrating either discovery phase or discovery accompanied by a validation phase. (H) Epidemiological data collected for depression study from various continents. (I) Percentage of males and females reported in depression studies.
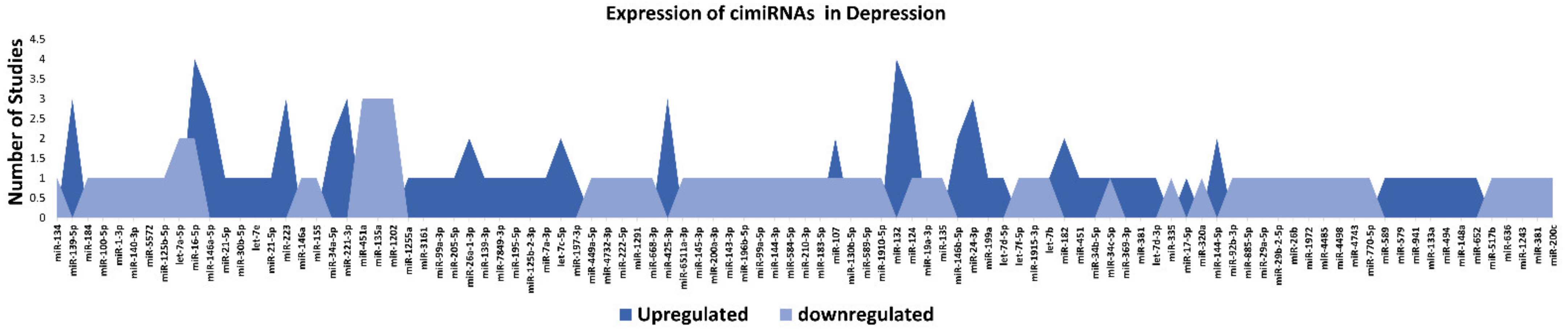
Figure 2. List and number of studies of dysregulated cimiRNAs in MDD.
miR-132 is one of the most enriched miRNAs in the brain that is indispensable for controlling axon growth, neuronal differentiation, migration, plasticity, etc. [21]. Lower expression of miR-132 suppresses BDNF expression, which results in MDD and suicidal behavior [22][23]. miR-124-3p is another significant neuronal miRNA involved in regulating neuronal fate determination, development, neuronal plasticity, etc. [24]. It is highly expressed in the brains of MDD patients [25][26] and is suspected to be a potential therapeutic target for designing novel anti-depressive drugs [27]. The Let-7 family of miRNAs are the most abundant miRNAs in the brain, and their dysregulated expression induces neuronal apoptosis, neuroinflammation, and depressive symptoms in humans [28]. To sum up, these investigations propose that specific dysregulation of brain miRNAs may predict some clinical manifestations observed in MDD patients.
Overall, the expression of 75% of cimiRNAs was found up-regulated in the body fluids of depressive patients (Figure 2). After ruling out other regulatory mechanisms such as transcription factor activity, histone modifications, methylation etc., it was speculated that most mRNA targets are lowered in MDD. In contrast, mRNA targets of downregulated cimiRNAs (25%) will have higher expression levels. Therefore, it is hypothesized that this disequilibrium of mRNA targets may disturb the balance of total gene expression in brain cells and contribute to the neuropsychiatry of MDD.
Qualitative analysis of all independent reports presented a limited overlap between them. A specific pattern of dysregulated cimiRNA depression signatures was formed after compiling all cimiRNAs from all 32 reports (Figure 2). Variable inconsistency was observed in these reports due to several factors, such as different methodologies adopted by various studies: diverse study designs, isolation, screening, and quantification techniques, etc. To begin, different subjective clinical scales (DSM-IV-V, HAM-D-HDRS, MADRS, HAMA, and CCMD-3) (Figure 1B) utilized by psychiatrists or physicians in these research studies may have impacted MDD diagnosis. MDD is assessed and diagnosed by predefined parameters that include three or more symptoms. However, this diagnostic approach does not seem appropriate because various patients presenting variable endophenotypes of MDD reported severe suicidal thoughts with just one or two defined symptoms. Furthermore, no prerequisite biological testing has been performed in primary clinical settings so far. Therefore, it is anticipated that it may influence the quality of results regarding the severity of MDD due to dependency on the specific parameters used in the MDD diagnosis. In most studies, body fluid samples from the control group (healthy individuals) and MDD patients for miRNA differential expression study were collected after 6–9 h fasting, whereas in some cases, no defined rule was followed. Similarly, sampling from the control group (healthy individuals) and patients was not of an appropriate match regarding age or sex in a few cases (Figure 1C). In addition, a small cohort size (less than 20 subjects) was a major issue due to bias that lowered the possibility of elucidation, either by statistical methods or dysregulated cimiRNAs (Figure 1D). Moreover, biofluid samples (blood, plasma, serum, and CSF) used in these studies contain differential cimiRNA contents (Figure 1D). For instance, CSF possesses more neuronal cimiRNAs than peripheral blood, whereas serum is enriched in more cimiRNAs than plasma due to the coagulation process. However, a comparison between CSF and serum or plasma neuronal cimiRNAs is also described in the results while studying genome-wide cimiRNA expression and adjusting Fisher’s value (p = 0.017) for different reports.
Techniques used for cimiRNA isolation are also another significant factor. Various characteristics of miRNAs such as small size, stable secondary structure, and very low counts in body fluids make their extraction and recovery quite challenging through extraction kits (Figure 1E). Different commercial kits possess different strengths (cons/pros) for cimiRNA isolation concerning yield and preference. Eleven different kits and protocols for cimiRNA extraction were described in 32 studies. Likewise, strategies for cimiRNA detection (Figure 1F) and isolation also influenced the quality of results. Real-time RT-PCR (qRT-PCR) is considered a gold standard method for specific and sensitive miRNA detection, but it was deployed to test a limited number of identified cimiRNAs, thus leaving a majority of dysregulated cimiRNAs undetected. In 21 studies, qRT-PCR was employed for direct cimiRNA quantification and detection, while four studies analyzed hundreds of cimiRNAs through chip microarray assay and qRT-PCR. In contrast, three studies used a real-time PCR-based array with pre-loaded PCR primers to broaden the comparison between samples. Furthermore, four studies reported next-generation sequencing (NGS), another promising method of qualitative and quantitative miRNA profiling. It identifies novel miRNAs but is often limited in terms of reproducibility and replicability due to technological and software issues. However, only one study screened cimiRNAs through miRNome sequencing and verified cimiRNAs by qRT-PCR.
The utility of dysregulated cimiRNAs was thoroughly investigated between discriminating subjects with MDD and healthy individuals in all 32 studies. Acute variations were found in diagnostic sensitivity and specificity, i.e., 21% of reports discovered dysregulated cimiRNAs together with MDD, whereas 79% of reports validated cimiRNAs after subsequent investigations (Figure 1G). Nine reports showed that cimiRNAs were analyzed by incorporation into receiver operating characteristic curves (ROCs) for their specific and sensitive roles as future biomarkers, whereas in other reports, cimiRNAs were merged into panels for more accuracy. Therefore, in terms of the present studies, one may predict that a panel of cross-validated cimiRNAs could serve as a future diagnostic marker.
Overall, the prevalence of MDD varies significantly across borders, affecting 6% of the population and increasing lifetime risk by three-fold (15–18%). It is estimated that one in five people go through at least one episode of depression at some stage in their lifetime. Epidemiological data from all 32 studies presented considerable disparity across countries (Figure 1H). Most of the subjects were reported from Asia, including 15 studies from China and individual studies from Taiwan, Iran, Turkey, and Israel. Six studies were reported from Europe (UK, Ireland, Romania, Sweden, Italy, and France), and six studies from North America (four reports from Canada and two from the USA). South America reported one study from Brazil, and Australia reported one study. Although these data include reports from most continents, they are still not enough to predict the exact consequences of MDD. For accurate diagnoses through cimiRNAs, a larger sample size with more populations in various regions should be analyzed.
Furthermore, the gender gap in the prevalence of MDD among males and females is another essential aspect. MDD prevails more frequently in females (usually two times higher) than males (Figure 1I). The data analyzed among male and female subjects present coherent results with other studies. However, this gender gap may be linked to various sex differences in susceptibility to various biological, psychological, and environmental factors involved in regulation on the micro and macro levels.
3. Profile of Dysregulated cimiRNA Targets in MDD
To discern biological processes and pathways influenced by dysregulated cimiRNAs, DAVID v6.8 was used to analyze the predicted mRNA targets of dysregulated cimiRNAs for KEGG pathways and GO categories (Figure 3). To avoid mistakenly identified cimiRNAs, cimiRNAs found in at least two studies with similar expression (20 out of 99 cimiRNAs, 20%: 4 downregulated, and 16 upregulated) were probed for analysis (Figure 2). However, the criteria for cimiRNA analysis were not made more strict (i.e., referring to cimiRNAs investigated in three or more reports), as this could have limited the number of cimiRNAs to less than 12% of the original cimiRNA list, which may not be suitable for analysis, as most of the studies were not NGS-based. mRNA targets were aligned by screening transcript–miRNA interactions with context plus score threshold less than −0.3, according to the TargetScan 7.2 algorithm. These mRNA targets complied with a list of 4489 transcripts that were further processed to analyze those mRNA transcripts marked by at least two dysregulated cimiRNAs. This second selection further restricted the list of mRNA transcripts to 632 species (14% of the original transcript list). This scrutiny was performed due to the following reasons: (i) most of the previous studies reporting overexpression/knockdown of single miRNAs did not present significant expression, whereas co-expression of two or more miRNAs targeting a specific mRNA ensured enhanced expression; (ii) most neuronal-specific mRNA transcripts are regulated by more than one miRNA collectively, improving mRNA transcript expression and targeting specific transcripts; (iii) false bioinformatic predictions were avoided, i.e., a stringent parameter was set to ensure that at least two cimiRNAs target each mRNAs species.
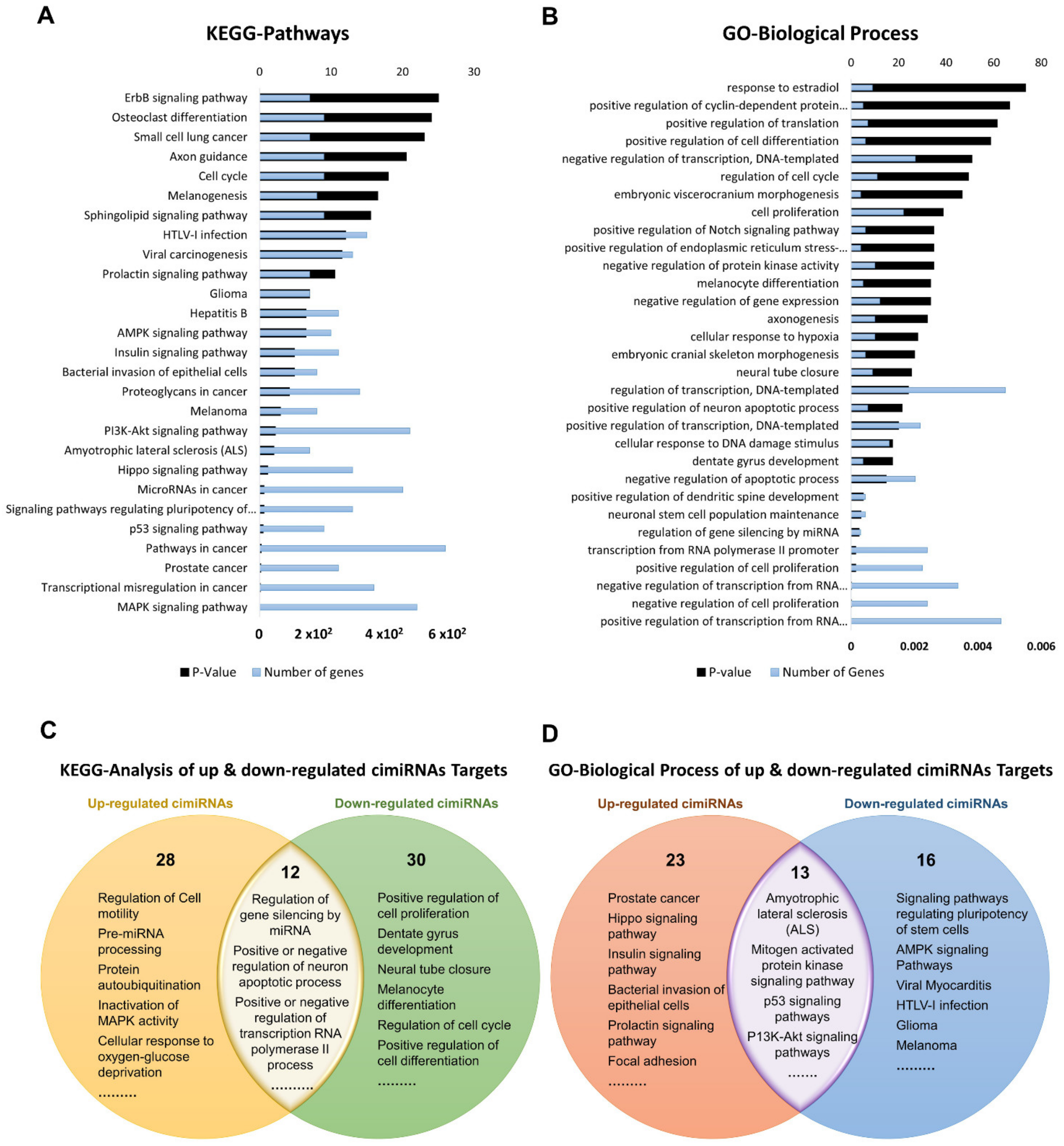
Figure 3. Functional annotation of dysregulated cimiRNA targets regulated by at least two dysregulated cimiRNAs identified in the studies. (A) Kyoto Encyclopedia of Gene and Genomes analysis for up- and downregulated targets of cimiRNAs. (B) Gene Ontology- Biological Process for up- and downregulated targets of cimiRNAs. (C) Venn diagram illustrating comparison of KEGG analysis for up- and downregulated cimiRNA target genes. (D) Venn diagram illustrating comparison of Gene Ontology-Biological Process for up- and downregulated cimiRNA target genes.
Dysregulated expression of cimiRNAs was confirmed twice in the MDD fluid samples with a specific transcript miRNA interaction score to ensure the chances of authentic expression in the MDD. Altogether, 27 KEGG pathways were found, significantly enriched by different dysregulated cimiRNAs (Figure 3A). Overlapped KEGG pathways of up- and down-regulated cimiRNA targets demonstrated various prominent processes such as amyotrophic lateral sclerosis (ALS), mitogen-activated protein kinase (MAPK) signaling pathway, p53 signaling pathways, and P13K-Akt signaling pathways along with other cancer-associated pathways, implicating their role in the pathophysiology of MDD (Figure 3C). Increasing evidence has suggested that MDD causes neuronal damage or neuronal death and is involved in the late onset of amyotrophic lateral sclerosis; for example, miR-34a [29] and miR-24 [30], targeting the BCL-2 (pro-apoptotic protein) are involved in lateral sclerosis, a rare neurological disease [31][32]. Interestingly, two cimiRNAs probed in current neuronal apoptosis could serve as future diagnostic markers.
Among various pathways, the MAPK signaling pathway plays a crucial role in various normal brain functions, including neuronal, synaptic, and structural plasticity, and is vital for neurotrophin/growth factor-mediated neuronal response [33]. This pathway is found in down-regulated cimiRNA (let-7a-5p) analysis as a significant pathway (p = 0.0038). Let-7a-5p downregulates RAS expression, suppressing the MAPK signaling pathway, and is positively associated with the depression-like phenotype [34][35]. P13K/Akt signaling cascade is strongly linked with the neurobiology of MDD [36]. Reduced AKT activity was found in several MDD patients [37]. Various cimiRNAs in the current study (miR-144, miR-137, let-7c, let-7b, miR-16, miR-182, miR-223, and miR-451) were found to regulate expression of several genes involved in the P13K/AKT pathway. Regarding MDD-associated mechanisms, lowered TP53 protein (tumor protein p53) expression increased anxiety and depressive-like behavior [38][39]. TP53 protein plays a pivotal role in the regulation of DNA repair, apoptosis, cell cycle progression, and brain development in the early stages. It is proposed to be a potential candidate for neurodegenerative and neuropsychiatric disorders [40][41]. Therefore, based on the current in-silico analysis, it is hypothesized that miR-26a-5p and let-7c targeting TP53 protein are involved in MDD-associated behaviors. Overall, the dysregulated KEGG pathways investigated by this analysis have been associated with MDD.
Gene ontology (GO) analyses (biological processes (GO-BP), cellular component (GO-CC) and molecular function (GO-MF)) of the dysregulated cimiRNA targets were performed. Most enriched biological processes (BP) (Figure 3B,D) were associated with ‘positive regulation of neuron apoptotic process’ (up miRNAs-7 targets, down miRNAs-5 targets), ‘negative regulation of neuron apoptotic process’ (up miRNAs-43 targets, down miRNAs-11 targets), ‘positive regulation of transcription RNA polymerase II process’ (up miRNAs-60 targets, down miRNAs-21 targets), ‘negative regulation of transcription RNA polymerase II process’ (up miRNAs- 26 targets, down miRNAs-11 targets), ‘axonogenesis’ (up miRNAs-10 targets, down miRNAs-5 targets), ‘cellular response to hypoxia’ (up miRNAs-10 targets, down miRNAs-5 targets) ‘regulation of gene silencing by miRNA’ (up miRNAs-4 targets, down miRNAs-3 targets), etc. Likewise, subcellular compartments (GO-CC) of up- and down-regulated cimiRNAs targets showed that the majority are localized in the nucleus (up miRNAs-190 targets, down miRNAs- 91 targets), nucleoplasm (up miRNAs-96 targets, down miRNAs-44 targets) and cytosol (up miRNAs-113 targets, down miRNAs- 50 targets) (Figure 4A). However, molecular functions (GO-MF) of their targets showed that DNA binding (up miRNAs-74 targets, down miRNAs-38 targets), zinc ion binding (up miRNAs-23 targets, down miRNAs-45 targets), transcription factor activity-specific sequence DNA binding activity (up miRNAs-51 targets, down miRNAs-23 targets), protein binding (up miRNAs-292 targets, down miRNAs-125 targets) and transcription factor binding (up miRNAs-51 targets, down miRNAs-23 targets) are the most enriched categories (Figure 4B). Altogether, the findings from this analysis are rather crucial, as a significant number of target transcripts are involved at transcriptional levels, depicting that cimiRNA dysregulation may disrupt mRNA transcription and involve a second wave of massive regulation, which includes wide aberrations at the transcription level.
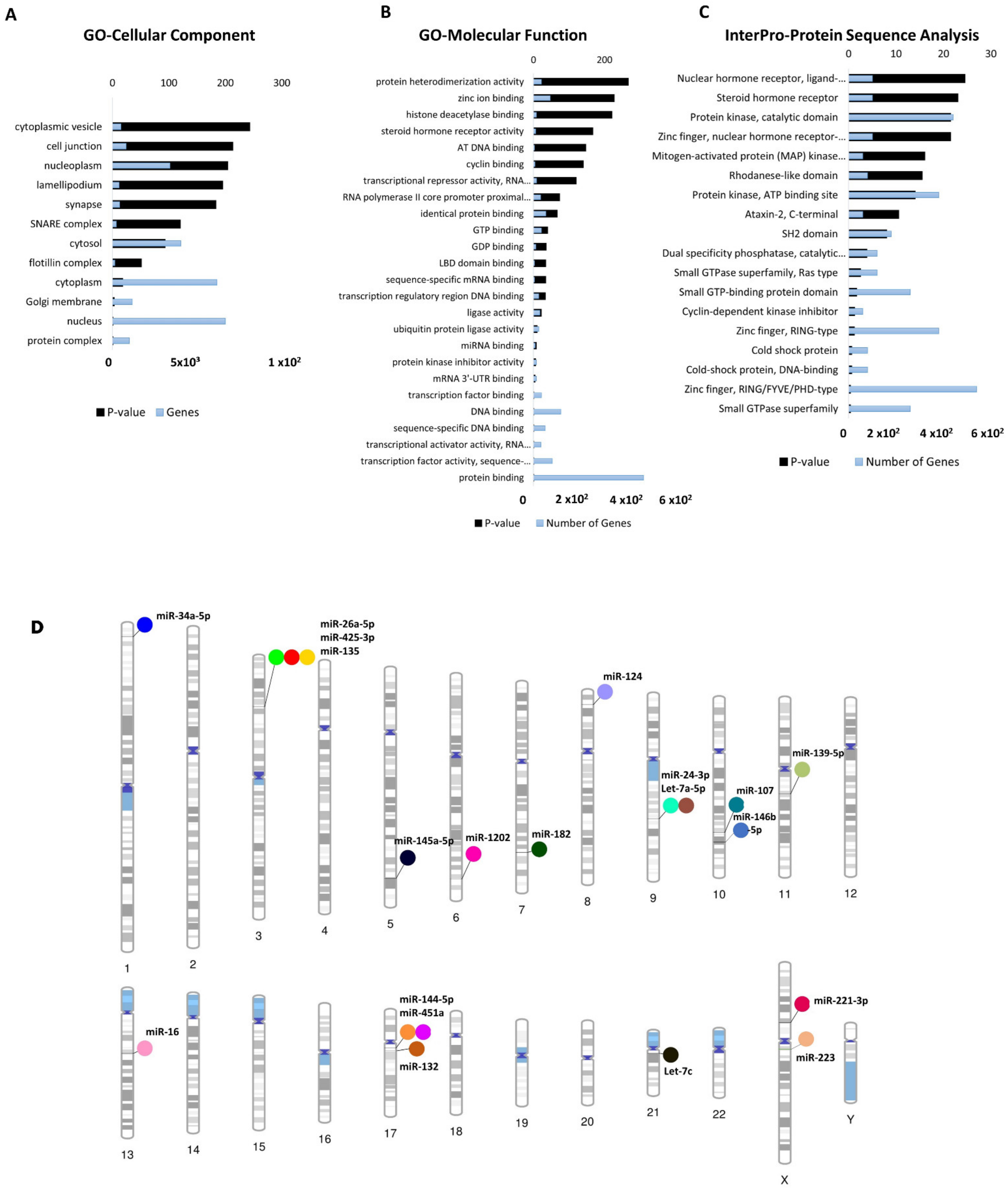
Figure 4. Functional annotation of dysregulated cimiRNA targets and chromosomal distribution of dysregulated cimiRNAs. (A) Gene Ontology-Cellular Components of up- and downregulated targets of cimiRNAs. (B) Gene Ontology-Molecular Functions of up- and downregulated targets of cimiRNAs. (C) InterPro-Protein sequence analysis of up- and downregulated targets of cimiRNAs. (D) Phenogram of the chromosomal distribution of dysregulated miRNAs in depression.
In addition, a predicted transcription factor (TF) mRNA target list was analyzed through the David InterPro protein sequence tool to garner a comprehensive understanding of transcription factors (TF) involved in the psychopathology of MDD. This analysis proposed Zinc finger domains (Znf) as the most prominent domains found in transcription factors involved in the MDD (Figure 4C). Literature survey showed that Znf domains are found in the Neur family of neuralized genes with neuralized homology repeat (NHR) domains [42][43]. Two Neur encoding proteins, specifically Neurl1 (neuralized E3 ubiquitin-protein ligase 1) and Neurl1B (neuralized E3 ubiquitin-protein ligase 1), are highly expressed in the brain and mediate various processes, including neuronal development and function through notch signaling [43][44][45][46]. A study on the adult mouse brain showed that disabling the Znf domain in mNeurl1 downregulated its expression, resulting in impaired hippocampal-dependent memory and synaptic plasticity [44]. Similarly, another study showed that mNeurl1-deficient mice manifested behavioral abnormalities such as ethanol hypersensitivity and olfactory discrimination [47].
Moreover, genome-wide association studies (GWAS) also presented the involvement of Neurl1B along with other genes in the development of various neuropsychiatric disorders [46][48][49], thus, suggesting that Znf domain dysfunction by these miRNAs may cause wide transcription changes that contribute to the neuropsychology of MDD.
Furthermore, MYCBP2, an E3-ubiquitin ligase MYC binding protein-2, was found to be targeted by miR-16, let-7c and miR-132 in this study. MYCBP2 is expressed in abundance in the brain [50][51] and modulates neuronal growth, synaptogenesis, and synaptic plasticity by mediating various signaling pathways, including p38 MAPK signaling cascades [50]. Alternatively, low expression of MYCBP2 resulted in elevated cAMP-synthesis, which causes neuronal excitability through PKA- and CREB-mediated aberrations in gene expression, thus leading to nociceptive behavior [52], which is associated with anxiety and depressive behavior in later stages [53][54]. Overall, this analysis shows that the mRNA transcripts of these cimiRNAs target various transcription factor proteins that play a significant role in the second wave of transcriptional changes in MDD progression and can be used as future diagnostic biomarkers.
4. Chromosomal Distribution of Dysregulated cimiRNAs in MDD
To shed more light, chromosomal coordinates of the 20 dysregulated cimiRNAs were obtained from miRbase (release 22.1) and Gene cards (The Human gene database) and visualized using Phenogram (Figure 4D). Surprisingly, a few cimiRNAs were clustered in close proximity, such as miR-26a-5p, miR135a, and miR-425-3p (3p22-p21), miR-24-3p and let 7a-5p (9q22.32), and miR-451a and miR-144-5p (17q11.2), suggesting that they might be co-regulated by specific transcription factors or methylation. A literature survey for potential vulnerabilities linked with these chromosomal regions revealed various gene expression changes that might accelerate depressive-like behaviors. For instance, two SNPs (-45 C/T and -196 G/A) were found in the CCK gene promoter region (3p22-p21) in patients with depressive and suicidal behavior [55]. This CCK encodes a neurotransmitter termed cholecystokinin, which is reported to be expressed at elevated levels in MDD and patients with suicidal intentions [56].
Similarly, the PTCH1 gene (9q22.32), which encodes the patched transmembrane protein, the principal receptor of Sonic hedgehog (Shh) signaling, plays a significant role in neuronal development and patterning hippocampus neurogenesis [57][58]. A study of heterozygous mice (Ptch1+/−) showed aberrant hippocampal morphogenesis with failed or altered neurogenesis. This dysregulated PTCH1 gene expression leads to phenotypic manifestations of neuropsychiatric disorders, including anxiety and MDD [59][60]. A significant number of genetic variants were observed in the promoter region, 5HHTLPR (5-hydroxytraptamine transporter-linked polymorphic region) of the SLC6A4 gene with cytogenic position 17q11.2 [61]. SLC6A4 encodes solute carrier family 6 member (serotonin transporter), whose abnormal expression levels are predominately associated with various episodes of depression [61]. The genetic mutations in 5HHTLPR are usually linked with the presence of “L allele” (16 GC-rich repeated elements of 20–30 bp) or absence of “S-allele” (14 repeated units of 20–30 bp except for deleted regions from 6th to 8th repeated elements) along with other rare variants containing 15, 19 and >20 repeats and SNPs [62][63][64][65]. Therefore, it is hypothesized that polymorphism or silencing of the SLC6A4 gene may result in abnormal expression of the SLC6A4 mRNA, which induces abnormal serotonin uptake and manifests mood disabilities [65][66]. Interestingly, through this analysis, we observed that cimiRNAs clustering on different chromosomes host only those protein-coding genes that are exclusively involved in MDD, thus identifying them as ideal cimiRNA biomarkers for monitoring the progression of MDD in future clinical trials.
5. Transcription Factors Regulating the Expression of Dysregulated cimiRNAs in MDD
Transcription factors regulating 20 cimiRNAs were investigated through the TransmiR v 2.0 experimental database to analyze TF sites 300 bp upstream and 100 bp downstream of each cimiRNA transcription site. Surprisingly, it was found that TF binding sites for hypoxia-inducible factor-1 (HIF-1), RE1 silencing transcription factor (REST), and T-cell acute lymphocytic leukemia protein 1 (TAL1) were found in most of the cimiRNAs, including cimiRNA clusters located at Chr-3 (miR-26a-5p, miR135a and miR-425-3p) and Chr-9 (miR-24-3p and let 7a-5p). Additionally, nuclear factor kappa B subunit 1 (NFKB1) and zinc finger E-box binding homeobox 1 (ZEB1) were also targeted by most of the cimiRNAs (Figure 5). Thus, to identify whether TFs with four or more miRNAs could be involved in MDD pathogenesis, transcriptomics expression data were thoroughly studied against their targets in MDD patients with healthy controls [67][68]. Accumulating shreds of evidence have shown that abnormal expression of HIF-1 and its target genes, including CCK, are profoundly associated with adult neurogenesis defects that trigger MDD pathophysiology [69][70][71]. Further, it is reported that transcriptional factor REST and its targets are also involved in the underlying pathology of MDD through deregulating neuronal function [71][72].
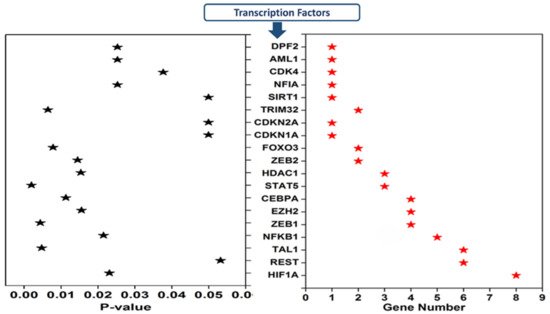
Figure 5. Transcription factors that regulate depression-associated cimiRNAs.
Moreover, Samaan et al., in 2015 reported that the obesity risk allele (rs2984618) in the TAL1 gene is strongly linked with a higher risk of MDD symptoms [72]. Therefore, in view of this analysis and the correlations of miRNAs and target genes with MDD, it is worth mentioning that somatic mosaicism in the genome of brain cells due to inherited genomic instability or epigenetic regulators, especially methylation, may not only deregulate miRNAs targeting transcription factors but also deregulate various genes that lead to the complex etiology of MDD. However, this domain has not yet been studied thoroughly and demands further investigation to understand the underlying neurobiology of MDD.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23031294
References
- Smith, K. Mental health: A world of depression. Nature 2014, 515, 180–181.
- Baxter, A.J.; Patton, G.; Scott, K.M.; Degenhardt, L.; Whiteford, H.A. Global Epidemiology of Mental Disorders: What Are We Missing? PLoS ONE 2013, 8, e65514.
- Gururajan, A.; Cryan, J.F.; Dinan, T.G. Molecular Biomarkers in Depression: Toward Personalized Psychiatric Treatment. In Personalized Psychiatry; Academic Press: Waltham, MA, USA, 2019; pp. 319–338.
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312.
- Dwivedi, Y. Emerging role of microRNAs in major depressive disorder: Diagnosis and therapeutic implications. Dialog. Clin. Neurosci. 2014, 16, 43–61.
- Wan, Y.; Liu, Y.; Wang, X.; Wu, J.; Liu, K.; Zhou, J.; Liu, L.; Zhang, C. Identification of Differential MicroRNAs in Cerebrospinal Fluid and Serum of Patients with Major Depressive Disorder. PLoS ONE 2015, 10, e0121975.
- Camkurt, M.A.; Acar, Ş.; Coşkun, S.; Güneş, M.; Güneş, S.; Yılmaz, M.F.; Görür, A.; Tamer, L. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J. Psychiatr. Res. 2015, 69, 67–71.
- Marí-Alexandre, J.; Sanchez-Izquierdo, D.; Gilabert-Estellés, J.; Barceló-Molina, M.; Braza-Boïls, A.; Sandoval, J. miRNAs Regulation and Its Role as Biomarkers in Endometriosis. Int. J. Mol. Sci. 2016, 17, 93.
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary MicroRNAs as Promising Biomarkers for Detection of Esophageal Cancer. PLoS ONE 2013, 8, e57502.
- Lipschitz, D.L.; Kuhn, R.; Kinney, A.Y.; Donaldson, G.W.; Nakamura, Y. Reduction in salivary α-amylase levels following a mind–body intervention in cancer survivors—An exploratory study. Psychoneuroendocrinology 2013, 38, 1521–1531.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Zheng, P.; Gao, H.C.; Li, Q.; Shao, W.H.; Zhang, M.L.; Cheng, K.; Yang, D.Y.; Fan, S.H.; Chen, L.; Fang, L.; et al. Plasma Metabonomics as a Novel Diagnostic Approach for Major Depressive Disorder. J. Proteome Res. 2012, 11, 1741–1748.
- Xu, H.-B.; Zhang, R.-F.; Luo, D.; Zhou, Y.; Wang, Y.; Fang, L.; Li, W.-J.; Mu, J.; Zhang, L.; Zhang, Y.; et al. Comparative proteomic analysis of plasma from major depressive patients: Identification of proteins associated with lipid metabolism and immunoregulation. Int. J. Neuropsychopharmacol. 2012, 15, 1413–1425.
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust MicroRNA Stability in Degraded RNA Preparations from Human Tissue and Cell Samples. Clin. Chem. 2010, 56, 998–1006.
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150.
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.P.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877.
- Balcells, I.; Cirera, S.; Busk, P.K. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol. 2011, 11, 70.
- Bartova, L.; Berger, A.; Pezawas, L. Is there a personalized medicine for mood disorders? Eur. Arch. Psychiatry Clin. Neurosci. 2010, 260, 121–126.
- Yuan, H.; Mischoulon, D.; Fava, M.; Otto, M.W. Circulating microRNAs as biomarkers for depression: Many candidates, few finalists. J. Affect. Disord. 2017, 233, 68–78.
- Van den Berg, M.M.J.; Krauskopf, J.; Ramaekers, J.G.; Kleinjans, J.C.S.; Prickaerts, J.; Briedé, J.J. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 2020, 185, 101732.
- Qian, Y.; Song, J.; Ouyang, Y.; Han, Q.; Chen, W.; Zhao, X.; Xie, Y.; Chen, Y.; Yuan, W.; Fan, C. Advances in Roles of miR-132 in the Nervous System. Front. Pharmacol. 2017, 8, 770.
- Xu, F.; Zheng, Z.; Zeng, Y.; Huang, H. MicroRNA-132 may play a role in coexistence of depression and cardiovascular disease: A hypothesis. Med. Sci. Monit. 2013, 19, 438–443.
- Yi, L.-T.; Li, J.; Liu, B.-B.; Luo, L.; Liu, Q.; Geng, D. BDNF–ERK–CREB signalling mediates the role of miR-132 in the regulation of the effects of oleanolic acid in male mice. J. Psychiatry Neurosci. 2014, 39, 348–359.
- Sun, Y.; Luo, Z.-M.; Guo, X.-M.; Su, D.-F.; Liu, X. An updated role of microRNA-124 in central nervous system disorders: A review. Front. Cell. Neurosci. 2015, 9, 193.
- Dwivedi, Y. microRNA-124: A putative therapeutic target and biomarker for major depression. Expert Opin. Ther. Targets 2017, 21, 653–656.
- Gu, Z.; Pan, J.; Chen, L. MiR-124 suppression in the prefrontal cortex reduces depression-like behavior in mice. Biosci. Rep. 2019, 39, BSR20190186.
- Roy, B.; Dunbar, M.; Shelton, R.C.; Dwivedi, Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology 2016, 42, 864–875.
- Gururajan, A.; Naughton, M.E.; Scott, K.A.; O’Connor, R.M.; Moloney, G.; Clarke, G.; Dowling, J.; Walsh, A.; Ismail, F.; Shorten, G.; et al. MicroRNAs as biomarkers for major depression: A role for let-7b and let-7c. Transl. Psychiatry 2016, 6, e862.
- Wang, X.; Liu, P.; Zhu, H.; Xu, Y.; Ma, C.; Dai, X.; Huang, L.; Liu, Y.; Zhang, L.; Qin, C. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res. Bull. 2009, 80, 268–273.
- Su, Y.; Wu, H.; Pavlosky, A.; Zou, L.-L.; Deng, X.; Zhang, Z.-X.; Jevnikar, A.M. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016, 7, e2333.
- Jia, J.; Le, W. Molecular network of neuronal autophagy in the pathophysiology and treatment of depression. Neurosci. Bull. 2015, 31, 427–434.
- Turner, M.R.; Goldacre, R.; Talbot, K.; Goldacre, M.J. Psychiatric disorders prior to amyotrophic lateral sclerosis. Ann. Neurol. 2016, 80, 935–938.
- Mahajan, G.J.; Vallender, E.J.; Garrett, M.R.; Challagundla, L.; Overholser, J.C.; Jurjus, G.; Dieter, L.; Syed, M.; Romero, D.G.; Benghuzzi, H.; et al. Altered neuro-inflammatory gene expression in hippocampus in major depressive disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 82, 177–186.
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the let-7 MicroRNA Family. Cell 2005, 120, 635–647.
- Dwivedi, Y. Brain-derived neurotrophic factor: Role in depression and suicide. Neuropsychiatr. Dis. Treat. 2009, 5, 433–449.
- Matsuda, S.; Ikeda, Y.; Murakami, M.; Nakagawa, Y.; Tsuji, A.; Kitagishi, Y. Roles of PI3K/AKT/GSK3 Pathway Involved in Psychiatric Illnesses. Diseases 2019, 7, 22.
- Karege, F.; Perroud, N.; Burkhardt, S.; Schwald, M.; Ballmann, E.; La Harpe, R.; Malafosse, A. Alteration in Kinase Activity But Not in Protein Levels of Protein Kinase B and Glycogen Synthase Kinase-3β in Ventral Prefrontal Cortex of Depressed Suicide Victims. Biol. Psychiatry 2007, 61, 240–245.
- Fan, T.; Hu, Y.; Xin, J.; Zhao, M.; Wang, J. Analyzing the genes and pathways related to major depressive disorder via a systems biology approach. Brain Behav. 2019, 10, e01502.
- Mahmood, S.; Evinova, A.; Škereňová, M.; Ondrejka, I.; Lehotský, J. Association of EGF, IGFBP-3 and TP53 Gene Polymorphisms with Major Depressive Disorder in Slovak Population. Central Eur. J. Public Health 2016, 24, 223–230.
- Levine, A.J. p53, the Cellular Gatekeeper for Growth and Division. Cell 1997, 88, 323–331.
- Komarova, E.A.; Chernov, M.V.; Franks, R.; Wang, K.; Armin, G.; Zelnick, C.R.; Chin, D.M.; Bacus, S.S.; Stark, G.R.; Gudkov, A.V. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity invivo. EMBO J. 1997, 16, 1391–1400.
- Ponting, C.P.; Mott, R.; Bork, P.; Copley, R.R. Novel Protein Domains and Repeats in Drosophila melanogaster: Insights into Structure, Function, and Evolution. Genome Res. 2001, 11, 1996–2008.
- Nakamura, H.; Yoshida, M.; Tsuiki, H.; Ito, K.; Ueno, M.; Nakao, M.; Oka, K.; Tada, M.; Kochi, M.; Kuratsu, J.-I.; et al. Identification of a human homolog of the Drosophila neuralized gene within the 10q25.1 malignant astrocytoma deletion region. Oncogene 1998, 16, 1009–1019.
- Pavlopoulos, E.; Trifilieff, P.; Chevaleyre, V.; Fioriti, L.; Zairis, S.; Pagano, A.; Malleret, G.; Kandel, E.R. Neuralized1 Activates CPEB3: A Function for Nonproteolytic Ubiquitin in Synaptic Plasticity and Memory Storage. Cell 2011, 147, 1369–1383.
- Song, R.; Koo, B.-K.; Yoon, K.-J.; Yoon, M.-J.; Yoo, K.-W.; Kim, H.-T.; Oh, H.-J.; Kim, Y.-Y.; Han, J.-K.; Kim, C.-H.; et al. Neuralized-2 Regulates a Notch Ligand in Cooperation with Mind Bomb-1. J. Biol. Chem. 2006, 281, 36391–36400.
- Lasky, J.L.; Wu, H. Notch Signaling, Brain Development, and Human Disease. Pediatr. Res. 2005, 57, 104–109.
- Ruan, Y.; Tecott, L.; Jiang, M.-M.; Jan, L.; Jan, Y.N. Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized. Proc. Natl. Acad. Sci. USA 2001, 98, 9907–9912.
- Hu, M.; Xia, Y.; Zong, X.; Sweeney, J.A.; Bishop, J.R.; Liao, Y.; Giase, G.; Li, B.; Rubin, L.H.; Wang, Y. Risperidone-induced changes in DNA methylation from peripheral blood in first-episode schizophrenia parallel neuroimaging and cognitive phenotype. bioRxiv 2020.
- Zhang, J.-P.; Aitchison, K.J.; Malhotra, A.K. The 12th annual pharmacogenetics in psychiatry meeting report. Psychiatr. Genet. 2014, 24, 218–220.
- Holland, S.; Coste, O.; Zhang, D.D.; Pierre, S.C.; Geisslinger, G.; Scholich, K. The Ubiquitin Ligase MYCBP2 Regulates Transient Receptor Potential Vanilloid Receptor 1 (TRPV1) Internalization through Inhibition of p38 MAPK Signaling. J. Biol. Chem. 2011, 286, 3671–3680.
- Yang, H.; Scholich, K.; Poser, S.; Storm, D.R.; Patel, T.B.; Goldowitz, D. Developmental expression of PAM (protein associated with MYC) in the rodent brain. Dev. Brain Res. 2002, 136, 35–42.
- Ehnert, C.; Tegeder, I.; Pierre, S.; Birod, K.; Nguyen, H.-V.; Schmidtko, A.; Geisslinger, G.; Scholich, K. Protein associated with Myc (PAM) is involved in spinal nociceptive processing. J. Neurochem. 2004, 88, 948–957.
- Seno, M.D.J.; de Assis, D.V.; Gouveia, F.V.; Antunes, G.F.; Kuroki, M.; Oliveira, C.C.; Santos, L.C.T.; Pagano, R.L.; Martinez, R.C.R. The critical role of amygdala subnuclei in nociceptive and depressive-like behaviors in peripheral neuropathy. Sci. Rep. 2018, 8, 13608.
- Gur, T.L. The Role of CREB Family Transcription Factors in Depression, Anxiety, and Antidepressant Response. Available online: https://repository.upenn.edu/dissertations/AAI3310377/ (accessed on 9 March 2020).
- Shindo, S.; Yoshioka, N. Polymorphisms of the cholecystokinin gene promoter region in suicide victims in Japan. Forensic Sci. Int. 2005, 150, 85–90.
- Jahangard, L.; Solgy, R.; Salehi, I.; Taheri, S.K.; Holsboer-Trachsler, E.; Haghighi, M.; Brand, S. Cholecystokinin (CCK) level is higher among first time suicide attempters than healthy controls, but is not associated with higher depression scores. Psychiatry Res. 2018, 266, 40–46.
- Pazzaglia, S.; Antonelli, F.; Casciati, A. Sonic hedgehog signaling controls dentate gyrus patterning and adult neurogenesis in the hippocampus. Neural Regen. Res. 2019, 14, 59–61.
- Hahn, H.; Wojnowski, L.; Zimmer, A.M.; Hall, J.; Miller, G.; Zimmer, A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 1998, 4, 619–622.
- Antonelli, F.; Casciati, A.; Tanori, M.; Tanno, B.; Linares, V.; Serra, N.; Belles, M.; Pannicelli, A.; Saran, A.; Pazzaglia, S. Alterations in Morphology and Adult Neurogenesis in the Dentate Gyrus of Patched1 Heterozygous Mice. Front. Mol. Neurosci. 2018, 11, 168.
- Winner, B.; Kohl, Z.; Gage, F.H. Neurodegenerative disease and adult neurogenesis. Eur. J. Neurosci. 2011, 33, 1139–1151.
- Ramamoorthy, S.; Bauman, A.L.; Moore, K.R.; Han, H.; Yang-Feng, T.; Chang, A.S.; Ganapathy, V.; Blakely, R.D. Antidepressant-and cocaine-sensitive human serotonin transporter: Molec-ular cloning, expression, and chromosomal localisation (paroxetine/biogenic amine uptake/placenta). Proc. Natl. Acad. Sci. USA 1993, 90, 2542–2546.
- Heils, A.; Teufel, A.; Petri, S.; Stöber, G.; Riederer, P.; Bengel, D.; Lesch, K.P. Allelic Variation of Human Serotonin Transporter Gene Expression. J. Neurochem. 1996, 66, 2621–2624.
- Lesch, K.-P.; Bengel, D.; Heils, A.; Sabol, S.Z.; Greenberg, B.D.; Petri, S.; Benjamin, J.; Müller, C.R.; Hamer, D.H.; Murphy, D.L. Association of Anxiety-Related Traits with a Polymorphism in the Serotonin Transporter Gene Regulatory Region. Science 1996, 274, 1527–1531.
- Heils, A.; Teufel, A.; Petri, S.; Seemann, M.; Bengel, D.; Balling, U.; Riederer, P.; Lesch, K.-P. Functional promoter and polyadenylation site mapping of the human serotonin (5-HT) transporter gene. J. Neural Transm. 1995, 102, 247–254.
- Shadrina, M.; Bondarenko, E.A.; Slominsky, P.A. Genetics Factors in Major Depression Disease. Front. Psychiatry 2018, 9, 334.
- Nakamura, M.; Ueno, S.; Sano, A.; Tanabe, H. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol. Psychiatry 2000, 5, 32–38.
- Li, S.; Li, Y.; Li, X.; Liu, J.; Huo, Y.; Wang, J.; Liu, Z.; Li, M.; Luo, X.-J. Regulatory mechanisms of major depressive disorder risk variants. Mol. Psychiatry 2020, 25, 1926–1945.
- Xu, F.; Yang, J.; Chen, J.; Wu, Q.; Gong, W.; Zhang, J.; Shao, W.; Mu, J.; Yang, D.; Yang, Y.; et al. Differential co-expression and regulation analyses reveal different mechanisms underlying major depressive disorder and subsyndromal symptomatic depression. BMC Bioinform. 2015, 16, 112.
- Li, G.; Zhao, M.; Cheng, X.; Zhao, T.; Feng, Z.; Zhao, Y.; Fan, M.; Zhu, L. FG-4592 Improves Depressive-Like Behaviors through HIF-1-Mediated Neurogenesis and Synapse Plasticity in Rats. Neurotherapeutics 2019, 17, 664–675.
- Malki, K.; Pain, O.; Tosto, M.G.; Du Rietz, E.; Carboni, L.; Schalkwyk, L. Identification of genes and gene pathways associated with major depressive disorder by integrative brain analysis of rat and human prefrontal cortex transcriptomes. Transl. Psychiatry 2015, 5, e519.
- Schmidt-Kastner, R.; van Os, J.; Esquivel, G.; Steinbusch, H.W.M.; Rutten, B. An environmental analysis of genes associated with schizophrenia: Hypoxia and vascular factors as interacting elements in the neurodevelopmental model. Mol. Psychiatry 2012, 17, 1194–1205.
- Otsuki, K.; Uchida, S.; Wakabayashi, Y.; Matsubara, T.; Hobara, T.; Funato, H.; Watanabe, Y. Aberrant REST-mediated transcriptional regulation in major depressive disorder. J. Psychiatr. Res. 2010, 44, 378–384.
This entry is offline, you can click here to edit this entry!
