Ca2+ is the most ubiquitous signaling molecule in the human body, regulating numerous biological functions including heartbeat, muscle contraction, neural function, cell development, and cell proliferation. Calcium dynamics result from fluxes in Ca2+ concentrations that vary in amplitude and duration, between intracellular compartments and between the intracellular and extracellular space and intracellular organelles. Calcium controls numerous biological processes by interacting with different classes of calcium binding proteins (CaBP’s), with different affinities, metal selectivities, kinetics, and calcium dependent conformational changes.
- rational design
- CaBP
- protein
- protein engineering
- synthesis
- fusion
1. Introduction
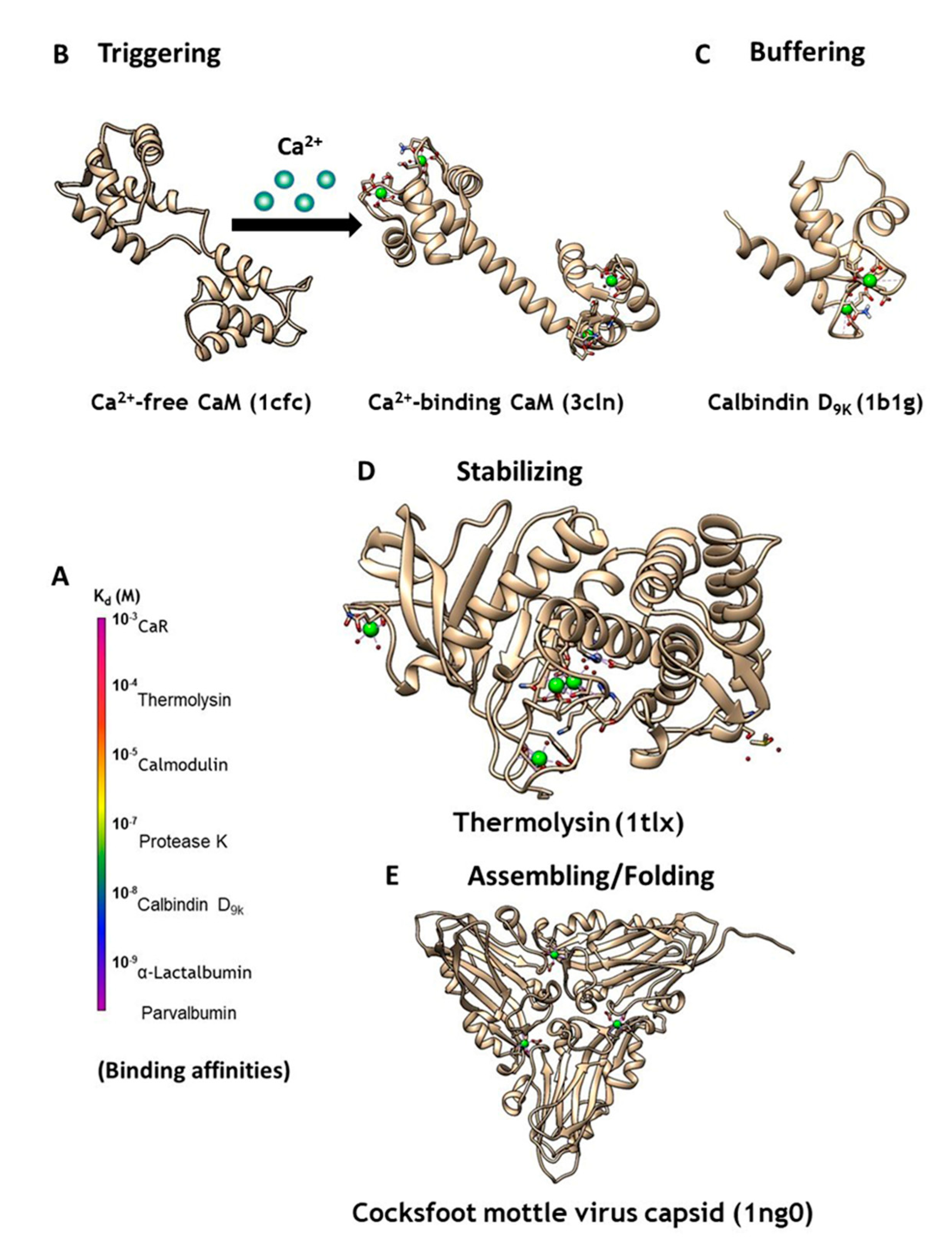
2. Classification and Prediction of Calcium Binding Sites in Proteins
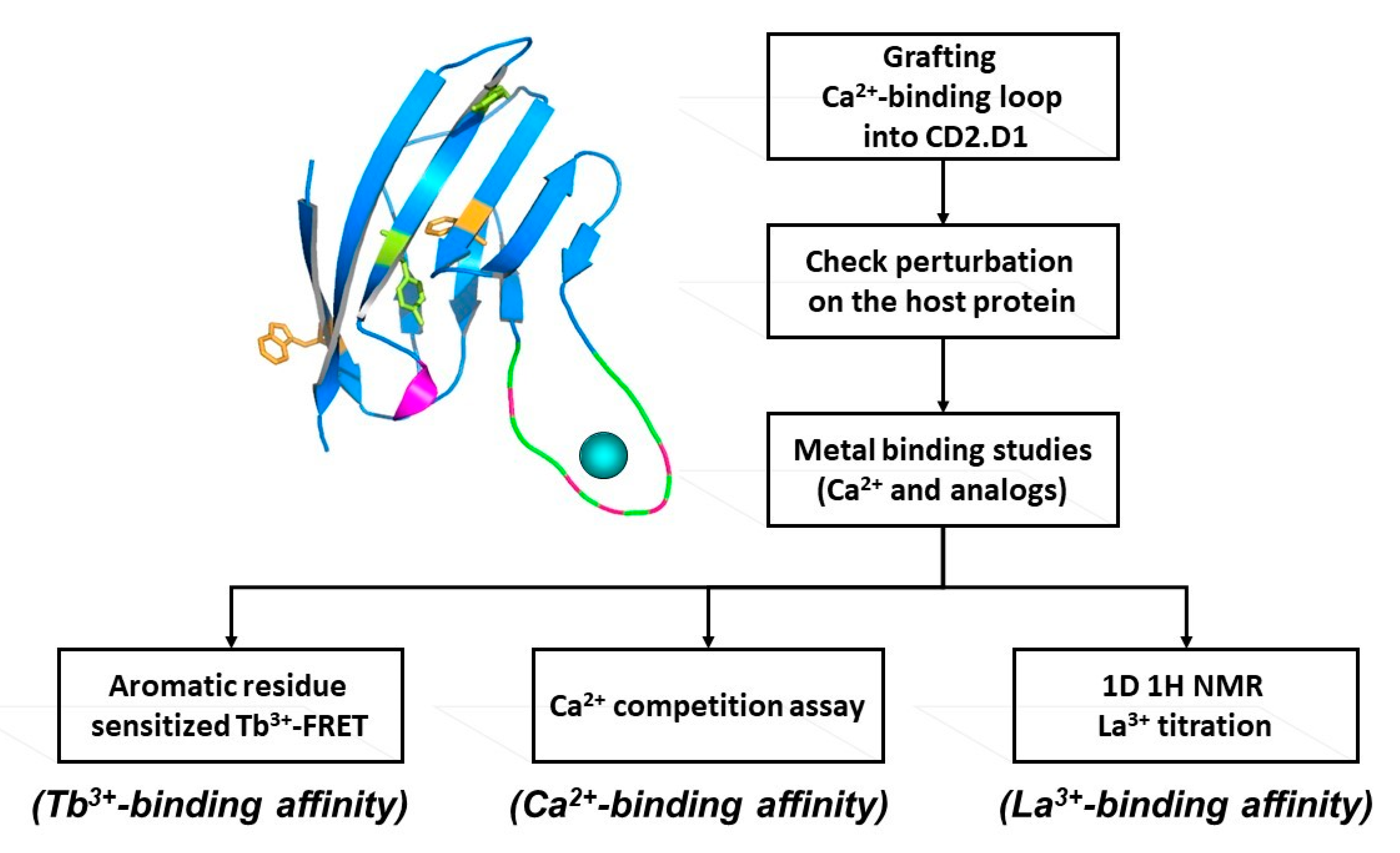
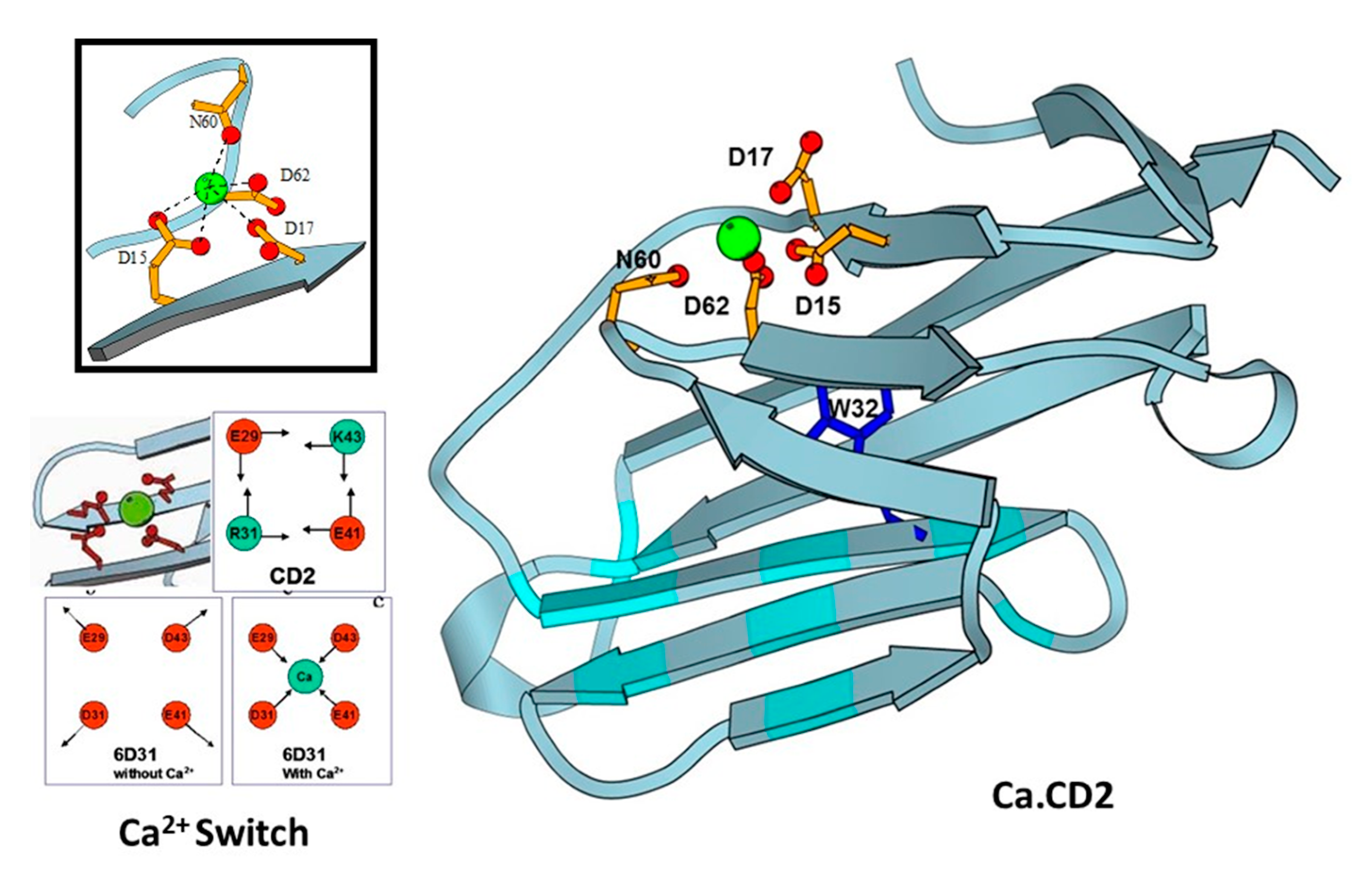
3. Development of Grafting and Design Approaches to Discover Key Factors that Contribute to Ca2+ Binding Affinities and Selectivities, and Ca2+-Induced Conformational Change
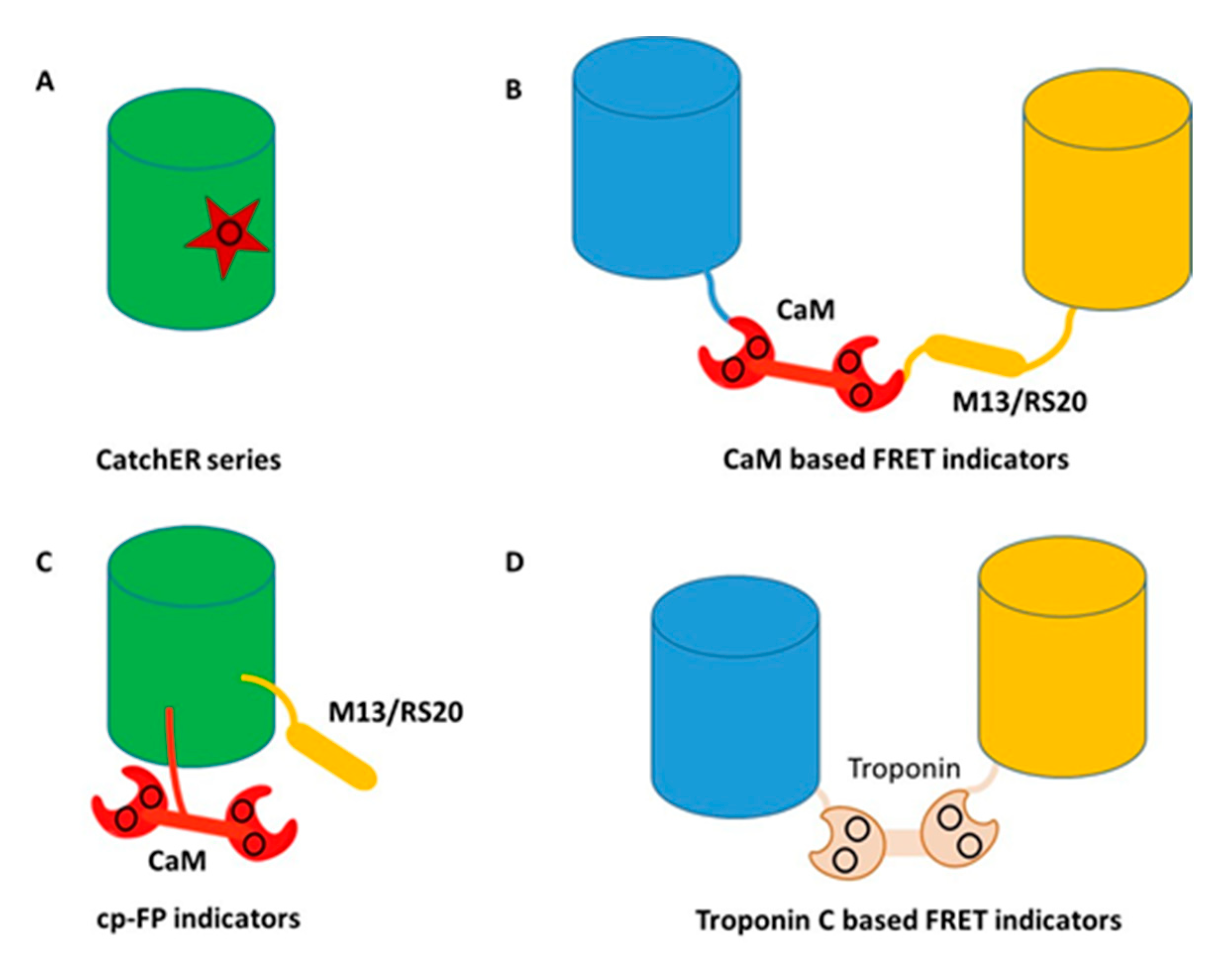
4. Development of Genetically Encoded Calcium Indicators (GECIs) Using Protein Engineering
5. Development of Calcium Sensors Using Rational Design
This entry is adapted from the peer-reviewed paper 10.3390/molecules25092148
References
- Miyawaki, A.; Llopis, J.; Heim, R.; Mccaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887.
- Rudolf, R.; Magalhaes, P.J.; Pozzan, T. Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytosolic camp dynamics in mouse skeletal muscle. J. Cell Biol. 2006, 173, 187–193.
- Bate, N.; Caves, R.E.; Skinner, S.P.; Goult, B.T.; Basran, J.; Mitcheson, J.S.; Vuister, G.W. A novel mechanism for calmodulin-dependent inactivation of transient receptor potential vanilloid 6. Biochemistry 2018, 57, 2611–2622.
- Zhu, M.X. Multiple roles of calmodulin and other Ca2+-binding proteins in the functional regulation of TRP channels. Pflug. Arch Eur. J. Physiol. 2005, 451, 105–115.
- Hofer, A.M.; Brown, E.M. Extracellular calcium sensing and signaling. Nat. Rev. Mol. Cell Biol. 2003, 4, 530–538.
- Zhang, C.; Miller, C.L.; Brown, E.M.; Yang, J.J. The calcium sensing receptor: From calcium sensing to signaling. Sci. China Life Sci. 2015, 58, 14–27.
- Zhang, C.; Zhang, T.; Zou, J.; Miller, C.L.; Gorkhali, R.; Yang, J.Y.; Schilmiller, A.; Wang, S.; Huang, K.; Brown, E.M.; et al. Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci. Adv. 2016, 2, E1600241.
- Jiang, Y.; Huang, Y.; Wong, H.C.; Zhou, Y.; Wang, X.; Yang, J.; Hall, R.A.; Brown, E.M.; Yang, J.J. Elucidation of a novel extracellular calcium-binding site on metabotropic glutamate receptor 1 (mglur1) that controls receptor activation. J. Biol. Chem. 2010, 285, 33463–33474.
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral calciomics: Interplays between Ca2+ and virus. Cell Calcium 2009, 46, 1–17.
- Kirberger, M.; Wang, X.; Deng, H.; Yang, W.; Chen, G.; Yang, J.J. Statistical analysis of structural characteristics of protein Ca2+-binding sites. J. Biol. Inorg. Chem. 2008, 13, 1169–1181.
- Kirberger, M.; Yang, J.J. Structural differences between Pb2+- and Ca2+-binding sites in proteins: Implications with respect to toxicity. J. Inorg. Biochem. 2008.
- Zhou, Y.; Yang, W.; Kirberger, M.; Lee, H.W.; Ayalasomayajula, G.; Yang, J.J. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins 2006, 65, 643–655.
- Deng, H.; Chen, G.; Yang, W.; Yang, J.J. Predicting Calcium-binding sites in proteins—A graph theory and geometry approach. Proteins 2006, 64, 34–42.
- Wang, X.; Kirberger, M.; Qiu, F.; Chen, G.; Yang, J.J. Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution. Proteins 2009, 75, 787–798.
- Wang, X.; Zhao, K.; Kirberger, M.; Wong, H.; Chen, G.; Yang, J. Analysis and prediction of calcium-binding pockets from apo protein structures exhibiting calcium-induced localized conformational changes. Protein Sci. 2010, 19, 1180–1190.
- Zhao, K.; Wang, X.; Wong, H.C.; Wohlhueter, R.; Kirberger, M.P.; Chen, G.; Yang, J.J. Predicting Ca2+-binding sites using refined carbon clusters. Proteins 2012, 80, 2666–2679.
- Yang, J.J.; Yang, J.; Wei, L.; Zurkiya, O.; Yang, W.; Li, S.; Zou, J.; Zhou, Y.; Maniccia, A.L.; Mao, H.; et al. Rational design of protein-based MRI contrast agents. J. Am. Chem. Soc. 2008, 130, 9260–9267.
- Zhou, Y.; Tzeng, W.P.; Yang, W.; Zhou, Y.; Ye, Y.; Lee, H.W.; Frey, T.K.; Yang, J. Identification of a Ca2+-binding domain in the rubella virus nonstructural protease. J. Virol. 2007, 81, 7517–7528.
- Huang, Y.; Zhou, Y.; Castiblanco, A.; Yang, W.; Brown, E.M.; Yang, J.J. Multiple Ca2+-binding sites in the extracellular domain of the Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 2009, 48, 388–398.
- Huang, Y.; Zhou, Y.; Yang, W.; Butters, R.; Lee, H.W.; Li, S.; Castiblanco, A.; Brown, E.M.; Yang, J.J. Identification and dissection of Ca2+-binding sites in the extracellular domain of Ca2+-sensing receptor. J. Biol. Chem. 2007, 282, 19000–19010.
- Ye, Y.; Lee, H.W.; Yang, W.; Shealy, S.; Yang, J.J. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J. Am. Chem. Soc. 2005, 127, 3743–3750.
- Ye, Y.; Shealy, S.; Lee, H.W.; Torshin, I.; Harrison, R.; Yang, J.J. A grafting approach to obtain site-specific metal-binding properties of EF-hand proteins. Protein Eng. 2003, 16, 429–434.
- Wilkins, A.L.; Ye, Y.; Yang, W.; Lee, H.W.; Liu, Z.R.; Yang, J.J. Metal-binding studies for a de novo designed calcium-binding protein. Protein Eng. 2002, 15, 571–574.
- Lee, H.W.; Yang, W.; Ye, Y.; Liu, Z.R.; Glushka, J.; Yang, J.J. Isolated Ef-loop III of calmodulin in a scaffold protein remains unpaired in solution using pulsed-field-gradient NMR spectroscopy. Biochim. Biophys. Acta 2002, 1598, 80–87.
- Franz, K.J.; Nitz, M.; Imperiali, B. Lanthanide-binding tags as versatile protein coexpression probes. Chembiochem 2003, 4, 265–271.
- Yang, W.; Jones, L.M.; Isley, L.; Ye, Y.; Lee, H.W.; Wilkins, A.; Liu, Z.R.; Hellinga, H.W.; Malchow, R.; Ghazi, M.; et al. Rational design of a calcium-binding protein. J. Am. Chem. Soc. 2003, 125, 6165–6171.
- Yang, W.; Wilkins, A.L.; Li, S.; Ye, Y.; Yang, J.J. The effects of Ca2+ binding on the dynamic properties of a designed Ca2+-binding protein. Biochemistry 2005, 44, 8267–8273.
- Yang, W.; Wilkins, A.L.; Ye, Y.; Liu, Z.R.; Li, S.Y.; Urbauer, J.L.; Hellinga, H.W.; Kearney, A.; Van Der Merwe, P.A.; Yang, J.J. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J. Am. Chem. Soc. 2005, 127, 2085–2093.
- Jones, L.M.; Yang, W.; Maniccia, A.W.; Harrison, A.; Van Der Merwe, P.A.; Yang, J.J. Rational design of a novel calcium-binding site adjacent to the ligand-binding site on CD2 increases its CD48 affinity. Protein Sci. 2008, 17, 439–449.
- Maniccia, A.W.; Yang, W.; Li, S.Y.; Johnson, J.A.; Yang, J.J. Using protein design to dissect the effect of charged residues on metal binding and protein stability. Biochemistry 2006, 45, 5848–5856.
- Ye, Y.; Lee, H.W.; Yang, W.; Yang, J.J. Calcium and lanthanide affinity of the Ef-loops from the C-terminal domain of calmodulin. J. Inorg. Biochem. 2005, 99, 1376–1383.
- Li, S.; Yang, W.; Maniccia, A.W.; Barrow, D., Jr.; Tjong, H.; Zhou, H.X.; Yang, J.J. Rational design of a conformation-switchable Ca2+- and Tb3+-binding protein without the use of multiple coupled metal-binding sites. FEBS J. 2008, 275, 5048–5061.
- Giepmans, B.N.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 2006, 312, 217–224.
- Lippincott-Schwartz, J.; Patterson, G.H. Development and use of fluorescent protein markers in living cells. Science 2003, 300, 87–91.
- Rizzuto, R.; Simpson, A.W.; Brini, M.; Pozzan, T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature 1992, 358, 325–327.
- Hofer, A.M.; Landolfi, B.; Debellis, L.; Pozzan, T.; Curci, S. Free dynamics measured in agonist-sensitive stores of single living intact cells: A new look at the refilling process. EMBO J. 1998, 17, 1986–1995.
- Persechini, A.; Lynch, J.A.; Romoser, V.A. Novel fluorescent indicator proteins for monitoring free intracellular Ca2+. Cell Calcium 1997, 22, 209–216.
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664.
- Palmer, A.E.; Tsien, R.Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 2006, 1, 1057–1065.
- Heim, N.; Griesbeck, O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 2004, 279, 14280–14286.
- Barykina, N.V.; Subach, O.M.; Piatkevich, K.D.; Jung, E.E.; Malyshev, A.Y.; Smirnov, I.V.; Bogorodskiy, A.O.; Borshchevskiy, V.I.; Varizhuk, A.M.; Pozmogova, G.E.; et al. Green fluorescent genetically encoded calcium indicator based on calmodulin/M13-peptide from fungi. PLoS ONE 2017, 12, E0183757.
- Tang, S.; Wong, H.C.; Wang, Z.M.; Huang, Y.; Zou, J.; Zhuo, Y.; Pennati, A.; Gadda, G.; Delbono, O.; Yang, J.J. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc. Natl. Acad. Sci. USA 2011, 108, 16265–16270.
- Reddish, F.N.; Miller, C.L.; Gorkhali, R.; Yang, J.J. Monitoring Er/Sr Calcium release with the targeted Ca2+ sensor catcher. J. Vis. Exp. 2017.
- Zhang, Y.; Reddish, F.; Tang, S.; Zhuo, Y.; Wang, Y.F.; Yang, J.J.; Weber, I.T. Structural basis for a hand-like site in the calcium sensor catcher with fast kinetics. Acta Crystallogr. D 2013, 69, 2309–2319.
- Zhuo, Y.; Solntsev, K.M.; Reddish, F.; Tang, S.; Yang, J.J. Effect of Ca2+ on the steady-state and time-resolved emission properties of the genetically encoded fluorescent sensor Catcher. J. Phys. Chem. B 2015, 119, 2103–2111.
- Zou, J.; Hofer, A.M.; Lurtz, M.M.; Gadda, G.; Ellis, A.L.; Chen, N.; Huang, Y.; Holder, A.; Ye, Y.; Louis, C.F.; et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry 2007, 46, 12275–12288.
