Avian orthoavulaviruses type-1 (AOaV-1) have transitioned from animal vaccine vector to a bona fide vaccine delivery vehicle in human. Owing to induction of robust innate and adaptive immune responses in mucus membranes in both birds and mammals, AOaVs offer an attractive vaccine against respiratory pathogens. The unique features of AOaVs include over 50 years of safety profile, stable expression of foreign genes, high infectivity rates in avian and mammalian hosts, broad host spectrum, limited possibility of recombination and lack of pre-existing immunity in humans. Additionally, AOaVs vectors allow the production of economical and high quantities of vaccine antigen in chicken embryonated eggs and several GMP-grade mammalian cell lines.
- avian orthoavulaviruses type-1
- human
- animal
- vaccines
1. Introduction
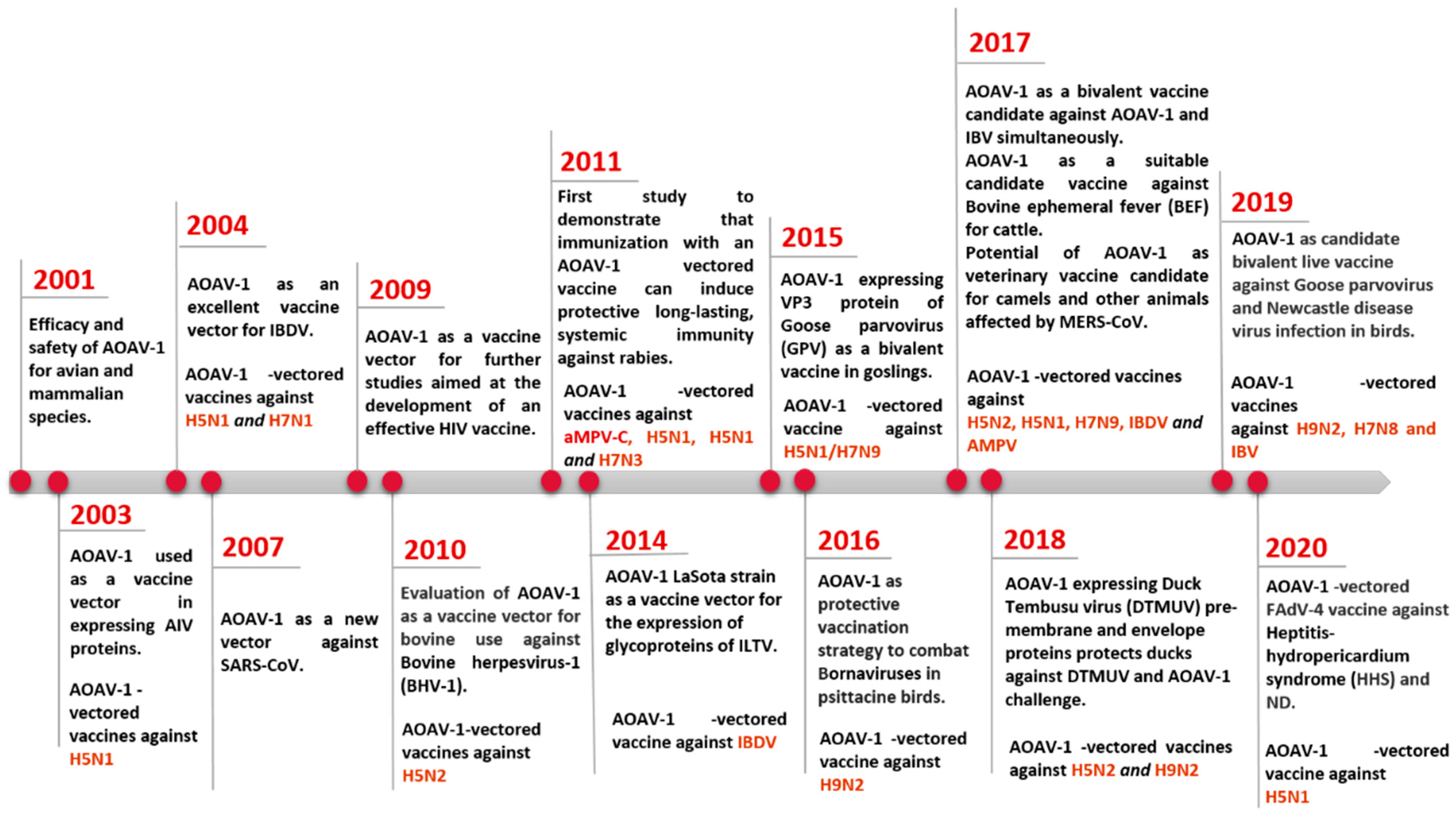
2. Genomic and Biological Features of Avian Orthoavulavirus Type-1
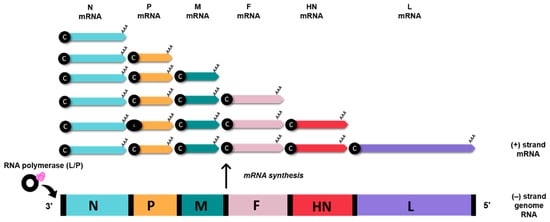
3. AOaV-1 as a Viral Vaccine Vector
4. Application of AOaV-1 as Vaccine Vector for Poultry and Animal Viruses
5. Current Application of AOaV-1 as Vaccine Vector for Emerging and Remerging Human Viruses
| Pathogen | AOaV-1 Backbone | Antigen | Insert Site | Animal Model | Vaccination (Route) | References |
|---|---|---|---|---|---|---|
| HIV-1 | Hitchner B1 | Gag | P/M | Mouse | i.n. | [6] |
| HIV-1 | La Sota | Gag | P/M | Mouse | i.n. | [36] |
| HIV-1 | La Sota | Gag; Env; Gag + Env | Env- P/M and Gag- HN/L; Gag- O/M and Env- HN/L; Env + Gag- P/M; Env- P/M; Gag- P/M | Guinea pigs/Mouse | i.n. | [34] |
| SIV | La Sota | gp160 | P/M | Guinea pigs/Mouse | i.n. | [37] |
| EBOV | Beaudette C and La Sota | GP | P/M | Rhesus monkeys | i.n/i.t. | [33] |
| EBOV | Chimeric AOaV-1 | GP | N/P, P/M, and M/F | Guinea pigs | i.n. | [38] |
| HPIV-3 | Beaudette C and La Sota | HN | P/M | African green monkeys and rhesus monkeys | i.n./i.t. | [3] |
| RSV | Hitchner B1 | F | P/M | Mouse | i.n. | [35] |
| Poliovirus | La Sota | P1 and 3CD | P1- P/M and 3CD- HN/L | Guinea pigs | i.n. | [39] |
| Lyme | LaSota/VF | BmpA + OspC | P/M | Hamsters | i.n./i.m./i.p. | [40] |
This entry is adapted from the peer-reviewed paper 10.3390/vaccines10020259
References
- Nakaya, T.; Cros, J.; Park, M.-S.; Nakaya, Y.; Zheng, H.; Sagrera, A.; Villar, E.; García-Sastre, A.; Palese, P. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 2001, 75, 11868–11873.
- Zhao, H.; Peeters, B.P.H. Recombinant Newcastle disease virus as a viral vector: Effect of genomic location of foreign gene on gene expression and virus replication. J. Gen. Virol. 2003, 84, 781–788.
- Bukreyev, A.; Huang, Z.; Yang, L.; Elankumaran, S.; St. Claire, M.; Murphy, B.R.; Samal, S.K.; Collins, P.L. Recombinant Newcastle disease virus expressing a foreign viral antigen is attenuated and highly immunogenic in primates. J. Virol. 2005, 79, 13275–13284.
- Hu, Z.; Ni, J.; Cao, Y.; Liu, X. Newcastle disease virus as a vaccine vector for 20 years: A focus on maternally derived antibody interference. Vaccines 2020, 8, 222.
- Choi, K.S. Newcastle disease virus vectored vaccines as bivalent or antigen delivery vaccines. Clin. Exp. Vaccine Res. 2017, 6, 72–82.
- Carnero, E.; Li, W.; Borderia, A.V.; Moltedo, B.; Moran, T.; García-Sastre, A. Optimization of human immunodeficiency virus gag expression by Newcastle disease virus vectors for the induction of potent immune responses. J. Virol. 2009, 83, 584–597.
- Tan, L.; Wen, G.; Qiu, X.; Yuan, Y.; Meng, C.; Sun, Y.; Liao, Y.; Song, C.; Liu, W.; Shi, Y.; et al. A recombinant la sota vaccine strain expressing multiple epitopes of infectious bronchitis virus (Ibv) protects specific pathogen-free (spf) chickens against ibv and ndv challenges. Vaccines 2019, 7, 170.
- DiNapoli, J.M.; Yang, L.; Suguitan, A.; Elankumaran, S.; Dorward, D.W.; Murphy, B.R.; Samal, S.K.; Collins, P.L.; Bukreyev, A. Immunization of primates with a Newcastle disease virus-vectored vaccine via the respiratory tract induces a high titer of serum neutralizing antibodies against highly pathogenic avian influenza virus. J. Virol. 2007, 81, 11560–11568.
- Kim, S.H.; Samal, S.K. Innovation in newcastle disease virus vectored avian influenza vaccines. Viruses 2019, 11, 300.
- Alexander, D.J. Newcastle disease. Br. Poultry Sci. 2001, 42, 5–22.
- Ganar, K.; Das, M.; Sinha, S.; Kumar, S. Newcastle disease virus: Current status and our understanding. Virus Res. 2014, 184, 71–81.
- Yusoff, K.; Tan, W.S. Newcastle disease virus: Macromolecules and opportunities. Avian Pathol. 2001, 30, 439–455.
- Acheson, N.H.; Kolakofsky, D.; Richardson, C. Paramyxoviruses and Rhabdoviruses. In Fundamentals of Molecular Virology; Acheson, N.H., Ed.; Wiley: Hoboken, NJ, USA, 2011; pp. 175–187.
- Panda, A.; Huang, Z.; Elankumaran, S.; Rockemann, D.D.; Samal, S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004, 36, 1–10.
- Iorio, R.M.; Mahon, P.J. Paramyxoviruses: Different receptors—Different mechanisms of fusion. Trends Microbiol. 2008, 16, 135–137.
- Samal, S.; Kumar, S.; Khattar, S.K.; Samal, S.K. A single amino acid change, Q114R, in the cleavage-site sequence of Newcastle disease virus fusion protein attenuates viral replication and pathogenicity. J. Gen. Virol. 2011, 92, 2333–2338.
- Krishnamurthy, S.; Huang, Z.; Samal, S.K. Recovery of a virulent strain of Newcastle disease virus from cloned cDNA: Expression of a foreign gene results in growth retardation and attenuation. Virology 2000, 278, 168–182.
- Bashir Bello, M.; Yusoff, K.; Ideris, A.; Hair-Bejo, M.; Hassan Jibril, A.; Peeters, B.P.H.; Omar, A.R. Exploring the prospects of engineered Newcastle disease virus in modern vaccinology. Viruses 2020, 12, 1–23.
- Huang, Z.; Elankumaran, S.; Panda, A.; Samal, S.K. Recombinant Newcastle disease virus as a vaccine vector. Poultry Sci. 2003, 82, 899–906.
- Peeters, B.P.H.; De Leeuw, O.S.; Verstegen, I.; Koch, G.; Gielkens, A.L.J. Generation of a recombinant chimeric Newcastle disease virus vaccine that allows serological differentiation between vaccinated and infected animals. Vaccine 2001, 19, 1616–1627.
- Huang, Z.; Krishnamurthy, S.; Panda, A.; Samal, S.K. High-level expression of a foreign gene from the most 3’-proximal locus of a recombinant Newcastle disease virus. J. Gen. Virol. 2001, 82, 1729–1736.
- Xiao, S.; Nayak, B.; Samuel, A.; Paldurai, A.; Kanabagattebasavarajappa, M.; Prajitno, T.Y.; Bharoto, E.E.; Collins, P.L.; Samal, S.K. Generation by reverse genetics of an effective, stable, live-attenuated Newcastle disease virus vaccine based on a currently circulating, highly virulent indonesian strain. PLoS ONE 2012, 7, e52751.
- Paldurai, A.; Kim, S.-H.; Nayak, B.; Xiao, S.; Shive, H.; Collins, P.L.; Samal, S.K. Evaluation of the contributions of individual viral genes to Newcastle disease virus virulence and pathogenesis. J. Virol. 2014, 88, 8579–8596.
- Rohaim, M.A.; Munir, M. A Scalable Topical Vectored Vaccine Candidate against SARS-CoV-2. Vaccines 2020, 8, 472.
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963.
- Pronker, E.S.; Weenen, T.C.; Commandeur, H.; Claassen, E.H.J.H.M.; Osterhaus, A.D.M.E. Risk in vaccine research and development quantified. PLoS ONE 2013, 8, e57755.
- Plotkin, S.; Robinson, J.M.; Cunningham, G.; Iqbal, R.; Larsen, S. The complexity and cost of vaccine manufacturing—An overview. Vaccine 2017, 35, 4064–4071.
- McLean, K.A.; Goldin, S.; Nannei, C.; Sparrow, E.; Torelli, G. The 2015 global production capacity of seasonal and pandemic influenza vaccine. Vaccine 2016, 34, 5410–5413.
- Gilbert, J.A. Seasonal and pandemic influenza: Global fatigue versus global preparedness. Lancet Respir. Med. 2018, 6, 94–95.
- Kim, S.H.; Samal, S.K. Newcastle disease virus as a vaccine vector for development of human and veterinary vaccines. Viruses 2016, 8, 183.
- Sun, W.; Leist, S.R.; McCroskery, S.; Liu, Y.; Slamanig, S.; Oliva, J.; Amanat, F.; Schäfer, A.; Dinnon, K.H., 3rd; García-Sastre, A.; et al. Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate. EBioMedicine 2020, 62, 103132.
- DiNapoli, J.M.; Kotelkin, A.; Yang, L.; Elankumaran, S.; Murphy, B.R.; Samal, S.K.; Collins, P.L.; Bukreyev, A. Newcastle disease virus, a host range-restricted virus, as a vaccine vector for intranasal immunization against emerging pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 9788–9793.
- DiNapoli, J.M.; Yang, L.; Samal, S.K.; Murphy, B.R.; Collins, P.L.; Bukreyev, A. Respiratory tract immunization of non-human primates with a Newcastle disease virus-vectored vaccine candidate against Ebola virus elicits a neutralizing antibody response. Vaccine 2010, 29, 17–25.
- Khattar, S.K.; Manoharan, V.; Bhattarai, B.; Labranche, C.C.; Montefiori, D.C.; Samal, S.K. Mucosal immunization with newcastle disease virus vector coexpressing HIV-1 env and gag proteins elicits potent serum, mucosal, and cellular immune responses that protect against vaccinia virus env and gag challenges. MBio 2015, 6, 1–10.
- Martinez-Sobrido, L.; Gitiban, N.; Fernandez-Sesma, A.; Cros, J.; Mertz, S.E.; Jewell, N.A.; Hammond, S.; Flano, E.; Durbin, R.K.; García-Sastre, A.; et al. Protection against respiratory syncytial virus by a recombinant Newcastle disease virus Vector. J. Virol. 2006, 80, 1130–1139.
- Maamary, J.; Array, F.; Gao, Q.; Garcia-Sastre, A.; Steinman, R.M.; Palese, P.; Nchinda, G. Newcastle disease virus expressing a dendritic cell-targeted HIV gag protein induces a potent gag-specific immune response in mice. J. Virol. 2011, 85, 2235–2246.
- Manoharan, V.K.; Khattar, S.K.; Labranche, C.C.; Montefiori, D.C.; Samal, S.K. Modified Newcastle Disease virus as an improved vaccine vector against Simian Immunodeficiency virus. Sci. Rep. 2018, 8, 8952.
- Yoshida, A.; Kim, S.H.; Manoharan, V.K.; Varghese, B.P.; Paldurai, A.; Samal, S.K. Novel avian paramyxovirus-based vaccine vectors expressing the Ebola virus glycoprotein elicit mucosal and humoral immune responses in guinea pigs. Sci. Rep. 2019, 9, 5520.
- Viktorova, E.G.; Khattar, S.K.; Kouiavskaia, D.; Laassri, M.; Zagorodnyaya, T.; Dragunsky, E.; Samal, S.; Chumakov, K.; Belov, G.A. Newcastle disease virus-based vectored vaccine against Poliomyelitis. J. Virol. 2018, 92, e00976-18.
- Xiao, S.; Kumar, M.; Yang, X.; Akkoyunlu, M.; Collins, P.L.; Samal, S.K.; Pal, U. A host-restricted viral vector for antigen-specific immunization against Lyme disease pathogen. Vaccine 2011, 29, 5294–5303.
