Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Coffee belongs to the genus Coffea of the family Rubiaceae and consists of over 100 species. The two main cultivated species, Coffea arabica L. (Arabica) and C. canephora Pierre ex A. Froehner (Robusta) accounted in 2020, on average, for about 60% and 40% of the world’s coffee production, respectively.
- coffee leaf rust
- rust variability
- effectors
- resistance
1. Introduction
Coffee is not only a vital presence in the daily life of a significant share of the world’s population, with consumption of over 3 billion cups of coffee a day, it is also the most valuable primary product in world trade, crucial to the economy of more than 50 countries, and constitutes the main source of livelihood for up to 25 million farmers and their families [1]. Traditionally, coffee has been viewed as a tropical commodity that links growing countries in the global South, along the so-called “bean belt” that lies between the tropics of Cancer and Capricorn, with consuming countries in the global North.
Coffee belongs to the genus Coffea of the family Rubiaceae and consists of over 100 species [2][3]. The two main cultivated species, Coffea arabica L. (Arabica) and C. canephora Pierre ex A. Froehner (Robusta) accounted in 2020, on average, for about 60% and 40% of the world’s coffee production, respectively [4]. Coffea liberica Bull ex Hiern (Liberian coffee), a third beverage species, is cultivated worldwide but is insignificant in terms of global trade [5].
Coffee leaf rust, caused by the biotrophic fungus Hemileia vastatrix Berkeley and Broome, is one of the main limiting factors of Arabica coffee production [6][7][8][9] causing production losses of over $1 billion annually worldwide [10].
Since the 19th century historical first burst of CLR, which caused the eradication of coffee cultivation in Ceylon (now Sri Lanka), the disease spread through coffee-growing countries of Asia and Africa until 1950. In the Americas, CLR was first detected in Brazil in 1970 and dispersed progressively across the coffee plantations of other Latin American countries [11][12][13]. In Hawaii, CLR was reported for the first time in 2020 [14]. Nowadays, CLR is present in all the coffee growing regions. Furthermore, in the last decade, the epidemic resurgence of CLR known as “the big rust” impacted several countries across Latin America and the Caribbean on a level similar to the one observed in Ceylon [15][16][17]. This epidemic is causing losses of several hundred of million dollars, with extremely serious social consequences in the agricultural sector. It has been associated with changes in climatic conditions, the ecology of coffee farms, and recurring economic shocks [15][16]. Initially, there was some speculation that the epidemic might have been caused by the emergence of new, more aggressive rust races, but this hypothesis has not yet been confirmed [16][18].
H. vastatrix infects the lower surface of the leaves where it produces chlorotic spots preceding the differentiation of suprastomatal, bouquet-shaped, orange-coloured uredinia, leading to premature defoliation (Figure 1).

Figure 1. Coffee leaf rust symptoms and signs. (A) Chlorotic spots and uredosporic sori on the lower leaf surface; (B) an uredosporic sorum observed under the scanning electron microscope; (C) defoliation in coffee plants as a result of disease (right side), contrasting with resistant plants in the field (Brazil). Photos taken by the authors.
According to Rhiney et al. [18], defoliation is CLR’s main effect, which reduces the plants’ photosynthetic activity affecting the quantity of fruits and can influence coffee quality.
Coffee-H. vastatrix interactions are governed by the Flor’s gene-for-gene relationship, and the resistance of coffee plants is conditioned at least by nine major dominant genes (SH1–SH9) singly or associated [11][19][20], although other major and minor genes may also be involved [11][21][22]. The use of coffee-resistant varieties is considered the most sustainable, efficient, and eco-friendly strategy to control this disease. Nevertheless, to improve the efficiency of the breeding process it is crucial to deepen our understanding of the complex mechanisms involved in the plant-pathogen interactions.
Plants have evolved highly complex defense mechanisms to protect themselves from various pathogens, with outcomes ranging from complete immunity, so called natural resistance or nonhost resistance, to susceptibility [23][24]. In addition to passive mechanisms, which include many preformed barriers such as waxy cuticles, rigid cell walls, and antimicrobial secondary metabolites [25], plants have evolved at least two lines of active defenses [26][27]. The first line provides basal defense against all potential pathogens and is based on the recognition of conserved microbe-associated or pathogen-associated molecular patterns (MAMPs/PAMPs) and host danger-associated molecular patterns (DAMPs), by pattern recognition receptors (PRRs) that activate PAMP-triggered immunity (PTI). Plants PRRs are either surface-localized receptor kinases (RKs) or receptor-like proteins (RLPs) containing various ligand-binding ectodomains that perceive PAMPs. Importantly, PTI has the potential to fend off multiple microbes, pathogenic or not, due to the conserved nature of PAMPs (e.g., bacterial flagellin, fungal chitin) across species, genera, family, or class. Thus, PRRs can provide resistance to most nonadapted pathogens, as well as contribute to basal immunity during infection [28][29][30]. The second layer of defense is activated when a given pathogen-derived molecule, called “effector”, is “specifically recognized” by plant receptor proteins encoded by R genes, resulting in effector-triggered immunity (ETI) that leads to fast and acute responses. Our understanding of ETI has evolved starting from the gene-for-gene theory [31], which describes the association between plants and their pathogens through the interaction of pathogen-derived avirulence (Avr) genes and plant-derived resistance (R) genes [26][32].
R genes mostly encode nucleotide-binding leucine-rich repeat receptor (NLRs) proteins. NLRs are modular proteins composed of several conserved domains that recognize effector proteins through different mechanisms. The direct recognition of the effectors by NLRs is described in the interaction between two flax NLRs and the rust Melampsora lini effectors [33]. In cases of indirect NLR-mediated effector recognition, either the NLR “guards” a host effector target and is activated upon detecting its molecular modification (the guard hypothesis), or a “sensor” NLR perceives the avirulent effector and activates a “helper” NLR that is responsible for triggering ETI [24][32].
Successful pathogens secrete effectors extracellularly or directly within the cytoplasm of the host cells, which can negatively interfere with PTI and ETI. As a result, plants are not able to defend themselves anymore and fall into the state of Effector-Triggered Susceptibility (ETS) [26][32].
Once activated, both PTI and ETI induce a downstream of similar defenses, such as rapid accumulation of reactive oxygen species (ROS), changes in cellular ion fluxes, activation of protein kinase cascades, production of stress-related hormones, cell wall modifications and changes in protein and gene expression [26][28][34]. These responses seem stronger and more prolonged during ETI when compared to PTI. ETI is typically associated with the hypersensitive reaction (HR), a form of programmed plant cell death localized at the infection sites [26][35]. HR is considered to be one of the most important factors in the restriction of the pathogen growth, particularly of obligate biotrophs [36][37] such as rust fungi, being responsible for race-specific resistance [38]. Despite their differences and particular features, the two-layered defenses (PTI and ETI) should be considered as a continuum resulting in the activation of an overlapping set of immune reactions [39].
Plants can also induce defense reactions to a broad range of pathogens due to prior exposure to pathogens or physical stress. This ability of plants to react to an invader by triggering local and systemic defense responses is known as systemic acquired resistance (SAR) [40]. It is considered one of the players in a multifaceted inducible defense system in plants, characterized by various signaling pathways and metabolic responses [41][42][43].
2. Coffee Leaf Rust (CLR) Causal Agent: Life Cycle and Biotrophic Infection Process
H. vastatrix (phylum Basidiomycota, class Pucciniomycetes, order Pucciniales) is a hemicyclic fungus producing urediniospores, teliospores and basidiospores, but only the dikaryotic urediniospores, which form the asexual part of the cycle, reinfect coffee leaves successively and are responsible for the disease [7]. H. vastatrix, like other rust fungi, is an obligate biotroph, which means that it can only feed, grow and reproduce on its living host(s). It differentiates specialized infection structures called haustoria that have been linked to pathogen nutrient uptake from the host cell. Haustoria also appear to play essential roles in plant-fungus recognition and delivery of secreted effector proteins into the host cytoplasm for the establishment of a successful biotrophic relationship [44][45][46][47].
The H. vastatrix infection process begins by the adhesion of urediniospores to the lower surface of the coffee leaves followed by its germination and appressoria differentiation over stomata. Adhesion may be regarded as a necessary prerequisite to establish the fungus on the leaf for successful infection [48]. After adhesion of rust urediniospores to the plant surface, the development of infection structures is the result of a sophisticated host surface recognition system. The tip of the dicaryotic germ tube is able to follow topographical features of the plant cuticle and thus increase the probability of encountering a stomatal opening [48]. Esterases seem to be involved in H. vastatrix urediniospores adhesion and appressoria differentiation [49].
The appressorium is the first infection structure from which H. vastatrix is able to penetrate the coffee leaf tissues, reaching sequentially the stages of penetration hypha, anchor and haustorial mother cell, which gives rise to a haustorium, that starts by infecting the stomatal subsidiary cells. Fungal growth continues with the formation of more intercellular hyphae, including HMCs and haustoria, in the spongy and palisade parenchyma, and even the upper epidermis. At this stage, it is possible to observe macroscopic chloroses on the leaf’s surface. Once the hyphae invade the substomatal cavities, they differentiate to form protosori, and about 3 weeks after the beginning of the infection urediniosporic sori (Figure 1) protrude through the stomata in a bouquet shape [7][20][50][51].
The development of the entire uredinial cycle depends on environmental conditions [7][18]. Urediniospore germination requires water and is optimal at about 24 °C; rain is the primary dispersal mechanism for spores. The time from initial infection to the production of a new sorus is shortened in higher temperatures. Thus, drier, cooler climates are not conducive for CLR spread, whereas warmer, humid climates favor an increase of urediospores (sporulation).
3. Rust Variability
The first indication of the adaptive capacity of H. vastatrix was the loss of resistance of C. liberica observed in Indonesia between 1880 and 1890, which contributed to the subsequent decline of Liberica coffee cultivation in that country [52]. The loss of resistance of Coorg Arabica variety in India alerted the occurrence of pathological shifts that led, from 1918 to 1920, to its replacement by Kent’s Arabica variety, which maintained its resistance to rust for several decades [53]. In 1932, Mayne, in India, presented the first experimental evidence of physiological specialization of H. vastatrix by differentiating two races using laboratory inoculations: one only virulent to Coorg (race 1) and the other to both Coorg and Kent’s (race 2) [54]. Afterwards, Mayne identified another two races with virulence to S.288 and S. 353 Arabica varieties [55].
Following the creation of the Coffee Rusts Research Center (CIFC, Centro de Investigação das Ferrugens do Cafeeiro) in Portugal in 1955, many samples of leaf rust and coffee genotypes were received from different regions of the world, assisting research of coffee resistance. Thanks to this work, the existence of Mayne’s races was confirmed at CIFC, and about 55 additional rust races have been discovered until today [7][11][20][21][55][56][57][58][59][60]. These races have been identified according to the virulence or avirulence reactions they induce when inoculated on a set of 27 coffee differentials: clonal lines of 5 C. arabica selections, 16 tetraploid hybrids of C. arabica × Coffea spp., and 6 Coffea spp. selections [7][11][21][59]. H. vastatrix races are attributed to isolates with distinct and unique combinations of virulence genes as inferred by Flor’s gene-to-gene theory and described as sequential Roman numerals in order of detection [19]. The race genotypes comprise virulence genes ranging from v1 to v9 in isolates derived from C. arabica and tetraploid interspecific hybrids, whereas those that attack diploid coffee species are not known due to the unavailability of genetic studies in these hosts [7][11][20]. The genetic characterization and confirmation of these virulence loci has not been possible so far, mainly because, on one side, no sexual phase in H. vastatrix’s life cycle has yet been identified, and on the other side, no direct link between phenotypic diversity and molecular diversity has been found. These limitations, together with the complexity and large size of H. vastatrix´s genome [61][62][63], have impaired further advances on the development of virulence diagnostic markers. Thus, the virulence profile characterization, particularly of isolates infecting Timor hybrid (HDT) derivatives, can only go as far as the available collection of coffee differential genotypes allows, leaving many genotypes incompletely identified [7][59]. For a long time, characterization of the population genetic variability in coffee rust was also intricated, as different studies reported different levels from low to high, but consistently no evidence of population structure could be found with respect to race, host or geographical origin [7]. Recent efforts in genomic research on coffee rust changed this paradigm, and for the first time three divergent genetic lineages were found, highly structured according to coffee hosts (C1 and C2, infecting diploid coffee species; and C3, infecting tetraploid coffee species), revealing footprints of introgression [64]. More recently, additional genomic data allowed further discovery of a well-resolved substructuring within the C3 lineage, which seems to follow a pattern of local adaptation [65]. With the likely increase of genomic resources in a short time, we might expect to improve our knowledge of population variability and virulence evolution and envision the future development of candidate diagnostic markers associated with rust pathotypes. Additionally, the first version of the core proteome of H. vastatrix urediniospores performed by nanoLC-MS/MS analysis has been presented [66]. Proteins were functionally annotated as being involved mainly in DNA integration, RNA-dependent DNA biosynthesis, proteolysis, translation, oxidation-reduction process, primary metabolism, nitrogen compound metabolism and macromolecule metabolic processes. The comparative analysis of the proteomes of the three rust races studied showed that 95% of the identified proteins (1780 out of 1874) were commonly detected among races. The identification of the protein factors (5%) that characterize each virulence profile is under analysis.
4. Mechanisms of Pathogenicity: The Search for H. vastatrix Effectors
The prediction of fungal effectors involved in plant interactions has been the focus of much attention by the research community working on plant diseases. Effectors are secreted proteins that move from the fungal to the host cells/tissues, where they induce or suppress plant defense responses since they modulate and interfere with the integrated plant immune system (including both PTI and ETI plant defense responses) [67]. Effector identification has the potential to speed up the selection of plant resistance genes and/or the removal of plant susceptibility genes in breeding programs of relevant agronomic varieties [68][69].
Fernandez et al. [70] obtained the first predict H. vastatrix’s secretome from C. arabica infected leaves by RNAseq. Two years later Talhinhas et al. [71], using RNAseq data from H. vastatrix in vitro structures (Figure 2), predicted the secretome of the prepenetration fungal structures. All together, these results generated a collection of 516 transcripts from H. vastarix that were predicted as putative secreted proteins. Surprisingly, a high number of these proteins were transcribed in prepenetration fungal structures, representing an early building of the pathogen’s virulence machine.
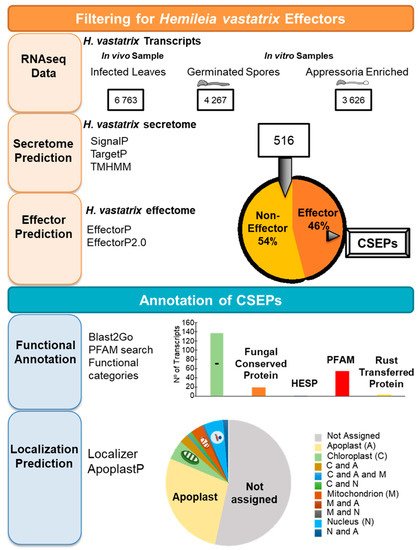
Figure 2. Pipeline for transcriptomic analysis of Hemileia vastatrix key differentiation stages, gene assembly, secretome prediction and bioinformatic annotation.
A machine learning approach was used to predict “Candidate secreted effector proteins” (CSEPs) from H. vastatrix secretome [72][73] (Figure 2). Almost half of the predicted secretome (516 transcripts) were also predicted to be CSEPs. In parallel, in silico annotation, such as the protein families database (Pfam) search, or functional annotation of those CSEPs sequences, revealed that 137 sequences do not contain any conserved protein domain. The activation of genes encoding putative effectors, namely the early activation of RTP (rust transferred proteins) and Gas1-orthologous genes, was observed in susceptible genotypes. Other putative effectors, (e.g., HESP—haustoria expressed secreted protein) seem to be specifically activated in the resistant genotypes and are preferentially expressed after the differentiation of the first haustoria [74][75]. The H. vastatrix CSEPs predicted subcellular localizations were apoplastic space (28%), nucleus (5%), chloroplast (5%), and mitochondrion (3%). However, for more than half of the CSEPs no localization was assigned (Figure 2).
The MEME algorithm was used to search for new conserved motifs amongst the sequences obtained by Fernandez et al. [70] and Talhinhas et al. [71]. Four putative secreted proteins were selected [76] (Figure 2) that shared some common features, such as gene organization and amino acid patterns, and their transcript level peaked seven days after inoculation in infected coffee leaves, when haustoria were the predominant fungal structure present. Together these results suggest a similar biological function or a common ancestor. The CSEPs were immunolocalized inside of intercellular hyphae and haustoria of H. vastatrix in infected leaves. The selected sequences showed the hallmark of true effectors, which needs to be supported by the ongoing effort of identifying the plant proteins interactors. Surprisingly, two CSEPs revealed splicing variants, which had a distinct transcription profile in leaves of Coffea spp. inoculated with different H. vastatrix isolates, and a different subcellular localization in leaves of Nicotiana benthamiana agroinfiltrated with the different variants. The splicing variants identified may contribute to increasing the “arsenal” diversity that H. vastatrix employs in the “arms race” against Coffea spp. immunity and may contribute to the diversification of H. vastatrix virulence potential [76].
The first CSEP from H. vastatrix to be identified as a bona fide effector was described in a noteworthy study conducted by Maia et al. [77] that started with the identification of CSEPs using RNAseq data. The novelty presented in this publication lies in the system used to deliver CSEPs into the cytoplasm of coffee leaf cells: the type-three secretion system of Pseudomonas syringae pv. garcae. Once proved that the CSEPs were inside the cells, the authors could determine which CSEPs influenced the growth of P. syringae pv. garcae in selected Coffea genotypes. They observed a suppression of bacterial growth by HVEC-016 effector, when inside of leaves from SH1 plants; suggesting that this effector can be recognized by a coffee plant with a SH1 background. This tool for delivering CSEPs inside leaf cells is of special relevance for further studies, considering the reduced number of molecular tools available for functional characterization in coffee plants.
This entry is adapted from the peer-reviewed paper 10.3390/agronomy12020326
References
- International Coffee Organization (ICO). Coffee Development Report 2019 Overview; International Coffee Organization: London, UK, 2019.
- Davis, A.P.; Tosh, J.; Ruch, N.; Fay, M. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of plastid and nuclear DNA sequences; implication for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linn. Soc. 2011, 167, 357–377.
- Charrier, A.; Berthaud, J. Botanical classification of coffee. In Coffee: Botany, Biochemistry and Production of Beans and Beverage; Clifford, M.N., Wilson, K.C., Eds.; Croom Helm: London, UK; Sydney, Australia, 1985; ISBN 978-1-4615-6659-5.
- International Coffee Organization (ICO). ICO Coffee Production 2020. Available online: https://www.ico.org/prices/po-production.pdf (accessed on 29 December 2021).
- Davis, A.P.; Chadburn, H.; Moat, J.; O’Sullivan, R.; Hargreaves, S.; Lughadha, E.N. High extinction risk for wild coffee species and implications for coffee sector sustainability. Sci. Adv. 2019, 5, eaav3473.
- van der Vossen, H.; Bertrand, B.; Charrier, A. Next generation variety development for sustainable production of arabica coffee (Coffea arabica L.): A review. Euphytica 2015, 204, 243–256.
- Talhinhas, P.; Batista, D.; Diniz, I.; Vieira, A.; Silva, D.N.; Loureiro, A.; Tavares, S.; Pereira, A.P.; Azinheira, H.G.; Guerra-Guimarães, L.; et al. Pathogen profile—The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Mol. Plant Pathol. 2017, 18, 1039–1051.
- Avelino, J.; Allinne, C.; Cerda, R.; Willocquet, L.; Savary, S. Multiple-disease system in coffee: From crop loss assessment to sustainable management. Annu. Rev. Phytopathol. 2018, 56, 611–635.
- Maghuly, F.; Jankowicz-Cieslak, J.; Bado, S. Improving coffee species for pathogen resistance. CAB Rev. 2020, 15, 1–18.
- Kahn, L.H. Quantitative framework for coffee leaf rust (Hemileia vastatrix), production and futures. Int. J. Agric. Ext. 2019, 7, 77–87.
- Bettencourt, A.J.; Rodrigues, C.J. Principles and practice of coffee breeding for resistance to rust and other diseases. In Coffee Agronomy; Clarke, R.J., Macrae, R., Eds.; Elsevier Applied Science Publishers LTD: London, UK; New York, NY, USA, 1988; Volume 4, pp. 199–234.
- McCook, S.; Peterson, P.D. The geopolitics of plant pathology: Frederick Wellman, coffee leaf rust, and cold war networks of science. Annu. Rev. Phytopathol. 2020, 58, 181–199.
- Wellman, F. Peligro de introducción de la Hemileia del café a las Américas. Turrialba 1952, 2, 47–50.
- Keith, L.; Sugiyama, L.; Brill, E.; Adams, B.-L.; Fukada, M.; Hoffman, K.; Ocenar, J.; Kawabata, A.; Kong, A.; McKemy, J.; et al. First report of coffee leaf rust caused by Hemileia vastatrix on coffee (Coffea arabica) in Hawaii. Plant Dis. 2021, 34, 2–4.
- Baker, P. The ‘Big Rust’: An update on the coffee leaf rust situation. Coffee Cocoa Int. 2014, 40, 37–39.
- Avelino, J.; Cristancho, M.; Georgiou, S.; Imbach, P.; Aguilar, L.; Bornemann, G.; Läderach, P.; Anzueto, F.; Hruska, A.J.; Morales, C. The coffee rust crises in Colombia and Central America (2008–2013): Impacts, plausible causes and proposed solutions. Food Secur. 2015, 7, 303–321.
- McCook, S.; Vandermeer, J. The Big Rust and the Red Queen: Long-term perspectives on coffee rust research. Phytopathology 2015, 105, 1164–1173.
- Rhiney, K.; Guido, Z.; Knudson, C.; Avelino, J.; Bacon, C.M.; Leclerc, G.; Aime, M.C.; Bebber, D.P. Epidemics and the future of coffee production. Proc. Natl. Acad. Sci. USA 2021, 118, e2023212118.
- Noronha-Wagner, M.; Bettencourt, A.J. Genetic study of the resistance of Coffea spp. to leaf rust. Can. J. Bot. 1967, 45, 2021–2031.
- Rodrigues, C.J.; Bettencourt, A.J.; Rijo, L. Races of the pathogen and resistance to coffee rust. Annu. Rev. Phytopathol. 1975, 13, 49–70.
- Várzea, V.M.P.; Marques, D.V. Population variability of Hemileia vastatrix vs. coffee durable resistance. In Durable Resistance to Coffee Leaf Rust; Zambolim, L., Zambolim, E., Várzea, V.M.P., Eds.; Universidade Federal de Viçosa: Viçosa, Brasil, 2005; pp. 53–74.
- De Almeida, D.P.; Samila, I.; Castro, L.; Antônio, T.; Mendes, D.O.; Alves, D.R. Receptor-Like Kinase (RLK) as a candidate gene conferring resistance to Hemileia vastatrix in coffee. Sci. Agric. 2021, 78, e20200023.
- Bettgenhaeuser, J.; Gilbert, B.; Ayliffe, M.; Moscou, M.J. Nonhost resistance to rust pathogens—A continuation of continua. Front. Plant Sci. 2014, 5, 664.
- Li, P.; Lu, Y.J.; Chen, H.; Day, B. The lifecycle of the plant immune system. Crit. Rev. Plant Sci. 2020, 39, 72–100.
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of plant defense system in response to microbial interactions. Front. Microbiol. 2020, 11, 1298.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Upson, J.L.; Zess, E.K.; Białas, A.; Wu, C.H.; Kamoun, S. The coming of age of EvoMPMI: Evolutionary molecular plant–microbe interactions across multiple timescales. Curr. Opin. Plant Biol. 2018, 44, 108–116.
- De Wit, P.J.G.M.; Mehrabi, R.; Van Den Burg, H.A.; Stergiopoulos, I. Fungal effector proteins: Past, present and future: Review. Mol. Plant Pathol. 2009, 10, 735–747.
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351.
- Han, G.Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83.
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–297.
- Lang, J.; Colcombet, J. Sustained incompatibility between MAPK signaling and pathogen effectors. Int. J. Mol. Sci. 2020, 21, 7954.
- Ravensdale, M.; Bernoux, M.; Ve, T.; Kobe, B.; Thrall, P.H.; Ellis, J.G.; Dodds, P.N. Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 2012, 8, e1003004.
- Peng, Y.; Van Wersch, R.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 2018, 31, 403–409.
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178.
- Heath, M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000, 44, 321–334.
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease resistance mechanisms in plants. Genes 2018, 9, 339.
- Periyannan, S.; Milne, R.J.; Figueroa, M.; Lagudah, E.S.; Dodds, P.N. An overview of genetic rust resistance: From broad to specific mechanisms. PLoS Pathog. 2017, 13, e1006380.
- Martins, D.; Araújo, S.D.; Rubiales, D.; Patto, M.C.V. Legume crops and biotrophic pathogen interactions: A continuous cross-talk of a multilayered array of defense mechanisms. Plants 2020, 9, 1460.
- Metraux, J. Systemic acquired resistance and salicylic acid: Current state of knowledge. Eur. J. Plant Pathol. 2001, 107, 13–18.
- Cavalcanti, F.R.; Resende, M.L.V.; Carvalho, C.P.S.; Silveira, J.A.G.; Oliveira, J.T.A. Induced defence responses and protective effects on tomato against Xanthomonas vesicatoria by an aqueous extract from Solanum lycocarpum infected with Crinipellis perniciosa. Biol. Control 2006, 39, 408–417.
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The “prime-ome”: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452.
- Gozzo, F.; Faoro, F. Systemic acquired resistance (50 years after discovery): Moving from the lab to the field. J. Agric. Food Chem. 2013, 61, 12473–12491.
- Heath, M.C. Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. 1997, 80, 713–720.
- Voegele, R.T.; Mendgen, K.W. Nutrient uptake in rust fungi: How sweet is parasitic life? Euphytica 2011, 179, 41–55.
- Catanzariti, A.M.; Dodds, P.N.; Lawrence, G.J.; Ayliffe, M.A.; Ellis, J.G. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 2006, 18, 243–256.
- Garnica, D.P.; Nemri, A.; Upadhyaya, N.M.; Rathjen, J.P.; Dodds, P.N. The ins and outs of rust haustoria. PLoS Pathog. 2014, 10, e1004329.
- Mendgen, K.; Voegele, R. Biology of rusts and mechanisms of infection. In Durable Resistance to Coffee Leave Rust; Zambolim, L., Zambolim, E.M., Várzea, V., Eds.; Universidade Federal de Viçosa: Viçosa, Brasil, 2005; pp. 233–248.
- Azinheira, H.G.; Guerra-Guimarães, L.; Silva, M.C.; Várzea, V.; Ricardo, C.P. Esterase activity and adhesion during the early stages of Hemileia vastatrix differentiation. In Proceedings of the 21st International Conference on Coffee Science (ASIC), France, Montpellier, 11–15 September 2006; pp. 1325–1329.
- Silva, M.C.; Nicole, M.; Rijo, L.; Geiger, J.P.; Rodrigues, C.J. Cytochemistry of plant- rust fungus interface during the compatible interaction Coffea arabica (cv. Caturra)-Hemileia vastatrix (race III). Int. J. Plant Sci. 1999, 160, 79–91.
- Silva, M.C.; Várzea, V.; Guerra-Guimarães, L.; Azinheira, H.G.; Fernandez, D.; Petitot, A.S.; Bertrand, B.; Lashermes, P.; Nicole, M. Coffee resistance to the main diseases: Leaf rust and coffee berry disease. Braz. J. Plant Physiol. 2006, 18, 119–147.
- Cramer, P.J.S. Review of literature of coffee research in Indonesia. Inter-Am. Inst. Agric. Sci. Misc. Publ. 1952, 15, 262.
- Narasimhaswamy, R.L. Coffee leaf disease (Hemileia) in India. Indian Coffee 1961, 25, 382–388.
- Mayne, W.W. Physiological specializalion of Hemileia vastatrix. B. & Br. Nature 1932, 129, 510.
- Mayne, W.W. Annual report of the coffee scientific officer 1938–39. Mys. Coffee Exp. Stn. 1939, 19, 1–16.
- Zhang, H.; Li, J.; Zhou, H.; Chen, Z.; Pereira, A.P.; Silva, M.C.; Várzea, V. Arabica coffee production in the Yunnan province of China. In Proceedings of the 24th International Conference on Coffee Science (ASIC), San José, Costa Rica, 12–16 November 2012; pp. 679–684.
- Noppakoonwong, U.; Khomarwut, C.; Hanthewee, M.; Jarintorn, S.; Hassarungsee, S.; Meesook, S.; Daoruang, C.; Nakai, P.; Lertwatanakiat, S.; Satayawut, K.; et al. Research and Development of Arabica Coffee in Thailand. In Proceedings of the 25th International Conference on Coffee Science (ASIC), Armenia, Colombia, 8–13 September 2014; pp. 42–49.
- Gichuru, E.K.; Ithiru, J.M.; Silva, M.C.; Pereira, A.P.; Várzea, V.M.P. Additional physiological races of coffee leaf rust (Hemileia vastatrix) identified in Kenya. Trop. Plant Pathol. 2012, 37, 424–427.
- Oliveira, B.; Bettencourt, A.J.; Rodrigues, C.J.; Rijo, L.; Azinheira, H.; Talhinhas, P.; Silva, M.C.; Várzea, V. Data Records of CIFC’s Research, 1955–2021. Unpublished work.
- Li, L.; Várzea, V.M.P.; Xia, Q.; Xiang, W.; Tang, T.; Zhu, M.; He, C.; Pereira, A.P.; Silva, M.C.; Wu, W.; et al. First report of Hemileia vastatrix (Coffee Leaf Rust) physiological races emergent in coffee germplasm collections in the coffee—Cropping regions of China. Plant Dis. 2021, 105, 4162.
- Tavares, S.; Ramos, A.P.; Pires, A.S.; Azinheira, H.G.; Caldeirinha, P.; Link, T.; Abranches, R.; Silva, M.D.; Voegele, R.T.; Loureiro, J.; et al. Genome size analyses of Pucciniales reveal the largest fungal genomes. Front. Plant Sci. 2014, 5, 422.
- Porto, B.N.; Caixeta, E.T.; Mathioni, S.M.; Vidigal, P.M.P.; Zambolim, L.; Zambolim, E.M.; Donofrio, N.; Polson, S.W.; Maia, T.A.; Chen, C.; et al. Genome sequencing and transcript analysis of Hemileia vastatrix reveal expression dynamics of candidate effectors dependent on host compatibility. PLoS ONE 2019, 14, e0215598.
- Cristancho, M.A.; Botero-Rozo, D.O.; Giraldo, W.; Tabima, J.; Riaño-Pachón, D.M.; Escobar, C.; Rozo, Y.; Rivera, L.F.; Durán, A.; Restrepo, S.; et al. Annotation of a hybrid partial genome of the coffee rust (Hemileia vastatrix) contributes to the gene repertoire catalog of the Pucciniales. Front. Plant Sci. 2014, 5, 594.
- Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol. Plant Pathol. 2018, 19, 1742–1753.
- Rodrigues, A.S.; Silva, D.N.; Várzea, V.; Paulo, O.S. Scaled-up genomic data uncovers a higher level of population genetic structuring in Hemileia vastatrix. In Proceedings of the 28th International Conference on Coffee Science (ASIC), Montpellier, France, 28 June–1 July 2021; p. 40.
- Guerra-Guimarães, L.; Pinheiro, C.; Leclercq, C.; Resende, M.L.V.; Jenny, J.; Ricardo, C.P.; Várzea, V. The first insight on the Hemileia vastatrix urediniospores proteome. In Proceedings of the 28th International Conference on Coffee Science (ASIC), Montpellier, France, 28 June–1 July 2021; p. 223.
- Boufleur, T.R.; Massola Júnior, N.S.; Tikami, Í.; Sukno, S.A.; Thon, M.R.; Baroncelli, R. Identification and comparison of Colletotrichum secreted effector candidates reveal two independent lineages pathogenic to soybean. Pathogens 2021, 10, 1520.
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant-Microbe Interact. 2014, 27, 196–206.
- Gorash, A.; Armonienė, R.; Kazan, K. Can effectoromics and loss-of-susceptibility be exploited for improving Fusarium head blight resistance in wheat? Crop J. 2021, 9, 1–16.
- Fernandez, D.; Tisserant, E.; Talhinhas, P.; Azinheira, H.; Vieira, A.; Petitot, A.S.; Loureiro, A.; Poulain, J.; da Silva, C.; Silva, M.C.; et al. 454-pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta-expressed pathogen-secreted proteins and plant functions in a late compatible plant-rust interaction. Mol. Plant Pathol. 2012, 13, 17–37.
- Talhinhas, P.; Azinheira, H.G.; Vieira, B.; Loureiro, A.; Tavares, S.; Batista, D.; Morin, E.; Petitot, A.-S.; Paulo, O.S.; Poulain, J.; et al. Overview of the functional virulent genome of the coffee leaf rust pathogen Hemileia vastatrix with an emphasis on early stages of infection. Front. Plant Sci. 2014, 5, 88.
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2016, 210, 743–761.
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110.
- Loureiro, A.; Tavares, S.; Azinheira, H.G.; Talhinhas, P.; Pereira, A.P.; Fernandez, D.; Silva, M.C. Identification and characterisation of Hemileia vastatrix effectors. In Proceedings of the 25th International Conference on Coffee Science (ASIC), Armenia, Colombia, 8–13 September 2014; p. 250.
- Silva, M.C.; Guerra-Guimarães, L.; Azinheira, H.G.; Talhinhas, P.; Tavares, S.; Loureiro, A.; Diniz, I.; Vieira, A.; Pereira, A.P.; Oliveira, H.; et al. Mechanisms involved in coffee—Hemileia vastatrix interactions: Plant and pathogen perspectives. In Proceedings of the 26th International Conference on Coffee Science (ASIC), Kunming, China, 13–19 November 2016; p. 14.
- Tavares, S.; Azinheira, H.G.; Voegele, R.; Thordal-Christensen, H.; Silva, M.; Talhinhas, P. Identification of Hemileia vastatrix candidate effectors reveals new ways of promoting pathogen variability through alternative splicing. In Proceedings of the 28th International Conference on Coffee Science (ASIC), Montpellier, France, 28 June–1 July 2021; p. 42.
- Maia, T.; Badel, J.L.; Marin-Ramirez, G.; Rocha, C.d.M.; Fernandes, M.B.; da Silva, J.C.F.; de Azevedo-Junior, G.M.; Brommonschenkel, S.H. The Hemileia vastatrix effector HvEC-016 suppresses bacterial blight symptoms in coffee genotypes with the SH1 rust resistance gene. New Phytol. 2017, 213, 1315–1329.
This entry is offline, you can click here to edit this entry!
