Knotted-like homeobox (KNOX) genes encode homeodomain-containing transcription factors (TFs) that modulate various important developmental processes in plants. While Class I KNOX TF genes are mainly expressed in the shoot apical meristems of both monocot and eudicot plants and are involved in meristem maintenance and/or formation, Class II KNOXTF genes exhibit diverse expression patterns and their precise functions have mostly remained unknown, until recently. The expression patterns of Class II KNOX TF genes in Arabidopsis, namely KNAT3, KNAT4, KNAT5, and KNAT7, suggest that TFs encoded by at least some of these genes, such as KNAT7 and KNAT3, may play a significant role in secondary cell wall formation.
- bioethanol
- KNOX II transcription factors
- saccharification
- secondary cell walls
- xylan
- xylem and fiber development
1. KNOX Genes and Encoded KNOX Proteins in Plants
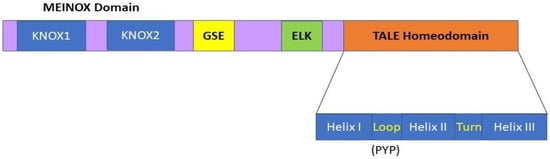
2. The Expression Patterns of Class II KNOX Genes in Plants Provide Some Clues about Their Functionality in SCW Formation
3. Genetic Mutations in Class II KNOX Genes Further Clarify Their Role in SCW Formation
| Target Gene | Mutation | Type of Mutation | Anatomy of Mutants | References |
|---|---|---|---|---|
| AtKNAT7 | irx11 | T-DNA insertion | Irregular xylem with collapsed vessels. | [30] |
| AtKNAT7 | - | Dominant repression | Reduced cell wall thickness of both xylem vessels and fibers; reduced composition of several monosaccharides from the cell walls. | [12] |
| AtKNAT7 | irx11 | Loss-of-function mutation | Thinner vessels walls resulted in a collapse of xylem vessels that showed the irx phenotype and thicker interfascicular fibers compared to controls; increase in lignin content. | [13] |
| AtKNAT3, AtKNAT4, AtKNAT5 | Single mutants | T-DNA insertion | No irx phenotype. | [15] |
| KNAT3/KNAT7 | Double mutant | T-DNA insertion | Enhanced irregular xylem (irx) phenotype characterized by weak inflorescence stem; reduced interfascicular fiber wall thickness and modified cell wall composition. | [15] |
| KNAT3/KNAT7 | Double mutant | Chimeric repression | Thinner interfascicular fiber cell walls compared to single mutants and wild type (WT); reduced cellulose and xylan and reduced S/G lignin ratio. | [16] |
| OsKNAT7 | CRISPR/CAS9 | T-DNA insertion | Thicker fiber cell walls; larger grain size due to cell expansion in spikelet bracts. | [35] |
| GhKNL1 | - | Dominant repression | Abnormal shorter fiber length. | [33] |
4. Targeted Genetic Manipulations in Class II KNOX Genes Confirm Their Role in SCW Formation
| Gene Used | Target Plant | Gene Modification Method | Impact on Transgenic Plants | References |
|---|---|---|---|---|
| AtKNAT7 | Arabidopsis | Overexpression | Thin interfascicular fiber walls, but no change in vessel wall thickness. | [13] |
| Cotton GhKNL1 | Arabidopsis | Overexpression | Thinner interfascicular fibers and slightly thinner vessel walls, but no change in xylary fibers. | [33] |
| Cotton GhKNAT7 | Arabidopsis | Overexpression | Reduced deposition of lignocellulose in interfascicular fibers, but no change in the SCWs of xylem fibers and vessels. | [24] |
| NbKNAT7 | Tobacco | Downregulation by VIGS and RNAi | Increased xylem proliferation with thin-walled fiber cells, increased polysaccharide extractability, and higher saccharification rate. | [14] |
| AtKNAT7 | Arabidopsis | Dominant repression | Reduced expression of SCW genes that resulted in thinner fiber cell walls with altered cell wall composition. | [12] |
| PtKNAT7 | Poplar | Overexpression | Enhanced expression of SCW genes, CesA8, IRX9, PAL, and CCR. | [17] |
| PtKNAT7 | Poplar | Downregulation by antisense | Reduced expression of SCW genes, reduced lignin content, altered lignin composition (S/G ratio), and increased saccharification. | [17] |
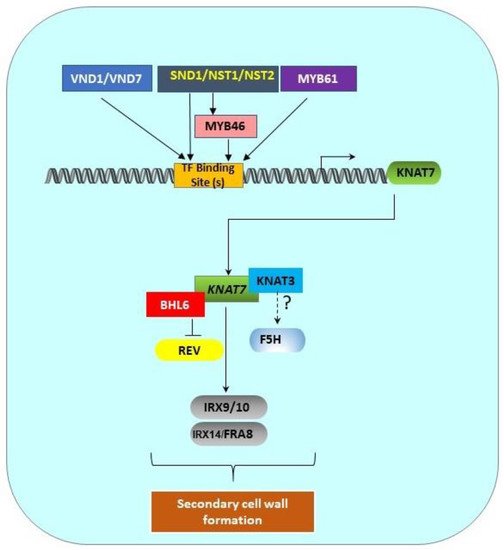
5. Transcriptional Network of the Class II KNOX Genes Involved in SCW Formation
This entry is adapted from the peer-reviewed paper 10.3390/plants11040493
