3. Fluorescence In Situ Hybridization Serves to Understand the Origin of Micronuclei
A breakthrough in the analyses of the localization of DNA damage at the chromosomal level in plants came with applying fluorescence in situ hybridization (FISH). It provides information on the possible ‘hot spots’ in plant genomes for DNA damage after the action of mutagens. Also, it gives information on the mechanisms of the biological effect of the individual agents that induce DNA damage. This knowledge is particularly crucial in plant mutagenesis, as the use of chemical and physical mutagens is the most common way to obtain mutants. This technique could detect even extremely small aberrations in dividing and non-dividing cells.
There is only one morphological type of micronuclei that may differ in size. The size of the micronucleus does not provide any information on whether it originated from chromosome fragments or entire chromosome(s), as the size may be related to the different degrees of the chromatin condensation. A more detailed analysis of the involvement of a specific chromosome or chromosome fragments in micronuclei formation is possible using fluorescence in situ hybridization (FISH). So far, FISH has not found such a wide application in the study of chromosome aberrations, including MN, in plants, as it has in humans [
36,
37,
38]. Different types of DNA probes for FISH are applied in plants, e.g., repetitive DNA sequences, single-locus chromosome-specific BAC clones, partial (e.g., arm), and whole chromosome paints. The limitations of the chromosome-specific DNA sequences in plants make the comprehensive identification of chromosome fragments in micronuclei using FISH still limited to a few species. Among the repetitive DNA sequences, centromere,
Arabidopsis thaliana (Arabidopsis)-type (TTTAGGG)n telomeric sequences, and ribosomal DNA (rDNA), which give strong and easily observed FISH signals, have found application in the detailed characterization of MN. These DNA sequences’ advantages are evolutionary conservation and location at a specific chromosome region. Repetitive dispersed DNA sequences are not a good source for probe pool for fluorescence in situ hybridization to study the origin of micronuclei.
FISH using 45S rDNA as the probe was first applied to localize the chromatin aberrations, such as translocations [
39] and anaphase bridges [
40], in
Arabidopsis thaliana. Applying the rDNA as probes showed rules regarding gamma-ray–induced MN formation in barley (
Hordeum vulgare) (
Figure 1).
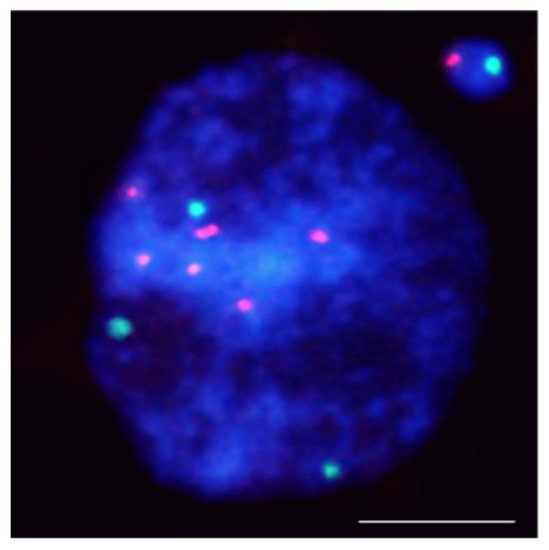
Figure 1. Hordeum vulgare interphase nuclei with the micronucleus induced by X-radiation. The nucleus was subjected to mcFISH with 5S rDNA (red) and 25S rDNA (green) probes. The micronucleus has one 5S rDNA and one 25S rDNA. Chromatin is stained with DAPI (blue). The bar represents = 10 µm.
5S rDNA-bearing chromosomes are shown to be more often involved in MN formation than NOR chromosomes in barley [
41,
42]. Similar rules regarding radiation-induced MN formation have been found in
Brachypodium distachyon [
43]. The hot spots for chromosome breakage in
Lolium multiflorum were not correlated with rDNA sites [
44].
The use of the centromere and telomere-specific DNA sequences for FISH also provided some rules regarding the origin of MN. It confirmed that the gamma ray-induced MN may originate from acentric fragments or whole lagging chromosomes. Thus, this approach allows the distinguishing of the micronuclei being a clastogenic and aneugenic effect of mutagens. However, most MN had only telomeric DNA signals, indicating that terminal deletion is the primary type of chromosome aberration leading to their formation (Figure 2).
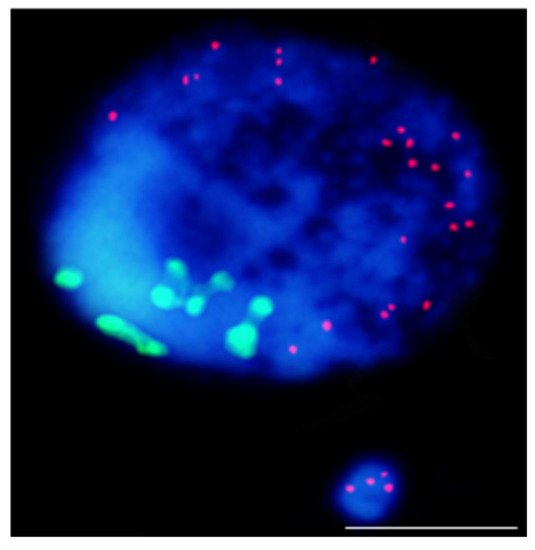
Figure 2. Results of mcFISH with telomeric (red) and centromeric (green) probes. Brachypodium distachyon interphase nuclei with micronucleus that were induced by X-radiation; micronucleus shows only telomeric DNA signals. The bar represents 5 µm. Micrograph by A. Kus.
Comparing the contribution of particular chromosome fragments in MN that are induced by different chemical clastogens, the maleic acid hydrazide (MH) and nitroso-N-methyl-urea (MNU) have shown the difference in the size of the chromosome fragments that are involved in the MN. Most MH-induced MN originated from large acentric fragments, whereas MNU-induced MN is from small terminal chromosome fragments [
41,
42].
FISH provides much more information about MN formation with DNA probes that are dedicated to different chromosomes or particular chromosomes. Standard A- and B-chromosome-specific probes were successfully used in the rye gamma-irradiated cells (
Secale cereale L.) [
45] for the detection of the translocations between the A- and B-chromosomes.
One of the FISH approaches that is used to detect and characterize micronuclei in plants is multicolor FISH (mcFISH). It is based on the two consecutive FISH experiments that use a pair or pairs of probe sets that are removed after each experiment and include the reprobing step. Combining more than two differently labeled DNA probes on the same nuclei slide makes this technique more informative [
46]. For the first time, mcFISH has been applied in human carcinogenicity studies [
47], then it has found application in mammalian cells [
48]. mcFISH is a common technique that is widely used in plants; however, it has narrow application in plant mutagenesis and genotoxicity. For the first time, this approach was applied in the analysis of the involvement of four different DNA sequences: 5S rDNA, 25S rDNA, the Arabidopsis-type (TTTAGGG)n telomeric sequence, and the Brachypodium-originated centromeric BAC clone CB33J12 in the micronuclei formation in
Brachypodium distachyon root-tip cells that were subjected to a chemical mutagen [
43].
The most advanced FISH-based approach in plants is chromosome painting (CP), which permits the selective visualization of entire chromosomes or their specific segments during mitosis as well the interphase [
49,
50,
51,
52,
53,
54]. The wide use of this technique for humans and mammals to determine the involvement of specific chromosomes in the formation of micronuclei showed that they preferentially comprise particular chromosomes that are related to the chromatin organization [
55]. The large amounts of repetitive DNA on all chromosomes are obstacles to CP on plants. CP is limited to a few plant species: Arabidopsis [
56], Brachypodium [
57], and few other species that are characterized by a small genome. mcFISH and CP with low repeat (small and large pools of bacterial artificial chromosomes (BAC)) clones that are specific for selected chromosomes, were applied to improve the ‘standard’ MN test in
Brachypodium distachyon (
Figure 3).
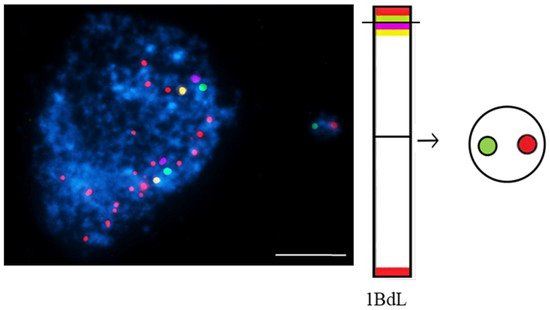
Figure 3. Brachypodium distachyon interphase nuclei with micronuclei that were induced by MH-treatment that were subjected to mcFISH with the following probes: telomeric sequence (red), I BAC pool (green), II BAC pool (violet), and III BAC pool (yellow). Chromatin is stained with DAPI (blue). The diagram next to the photomicrographs shows the putative origins of the micronuclei. Transverse dashed lines indicate chromosome breakpoint. The scale bar = 5 µm. Micrographs by A. Kus.
BAC-FISH-based chromosome painting provides new information on the composition, origin, and mechanisms of micronuclei formation that is induced by MH-treatment and X-radiation in Brachypodium by showing the ‘fragile spots’ of DNA breaks [
58]. Site-specific DNA breaks in chromosomes Bd4 and Bd5 were shown [
59].
To summarize, FISH provides new insights into the localization of DNA breaks on plant chromosomes, proving the non-random distributions of chromosome aberrations. The reasons for this non-random distribution may be the spatial organization of the nucleus at the interphase, the diverse transcriptional activity of specific chromosome regions, and chromosome size. Single BAC-FISH-based chromosome barcoding and ‘chromosome painting’ approaches have proven to be effective in analyzing the mechanism of micronuclei formation in plants after mutagenic treatment. The advantages of the FISH technique in terms of accuracy and quality of quantitative analyses make the technique one that is likely to become more widespread in DNA damage studies in plants.