Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Andrology
The role of Dickkopf-3 (Dkk3)/REIC (The Reduced Expression in Immortalized Cells), a Wnt-signaling inhibitor, in male reproductive physiology remains unknown thus far.
- Dkk3/REIC
- fibrous sheath
- knock-out
- RNA-seq
1. Introduction
Infertility is a serious problem that affects over 14% of couples in the world, and half of the cases can be attributed to male factors [1]. With the help of modern techniques in molecular biology, genetic disorders have been proven to be a major cause of male infertility, especially for those with non-obstruction in the genital ducts [2]. To date, over 2000 genes have been reported to participate in male reproductive physiology, and new findings continue emerging [3]. According to recent studies, Wnt signaling, a classical intercellular pathway that regulates cellular proliferation and tissue development, plays a pivotal role in male reproductive function [4,5].
Among the Wnt signal community, Dickkopf (Dkk) family, a negative regulator of Wnt signaling, has recently attracted attention from scholars in male reproduction fields. Currently, five members of the family have been found, which are known as Dkk1-4 and Dkkl1 (Dickkopf-like protein 1) [6]. For years, our institution has been dedicated to the research of Dkk3 and first identified its human homologous counterpart as a gene whose expression is reduced in immortalized cells, thus, also named Dkk3 as REIC (The Reduced Expression in Immortalized Cells) [7].
Dkk3/REIC shares high sequence similarity with Dkkl1, and they are believed to arise from the same ancestral precursor [6]. There are several studies reporting that Dkkl1 is deeply involved in the male reproductive process, as overexpressing Dkkl1 in male mice can severely impair their spermatogenesis, and sperm from Dkkl1−/− mice exhibit a decreased sperm-egg binding capacity [8,9].
2. Spermiation Failuare and Malposition of Differentiated Spermatids Exist in Testicular Sections of Dkk3/REIC-KO Mice
To investigate morphological changes of spermatogenesis after Dkk3/REIC was knocked out, the PAS-stained testicular sections of Dkk3/REIC-KO and Dkk3/REIC-WT mouse were examined. We noticed that spermiation failure existed in Dkk3/REIC-KO mice testes, and disordered placement of differentiated spermatids (which was most clearly displayed in stage VIII–IX) was commonly found in their testicular seminiferous tubles. As shown in Figure 1, differentiated spermatids were absent from the centers of the tubules and irregularly wandered adjacent to the basement membrane. However, the average amount of testiclular germ cells revealed no significant difference between the Dkk3/REIC-WT and Dkk3/REIC-KO mice.
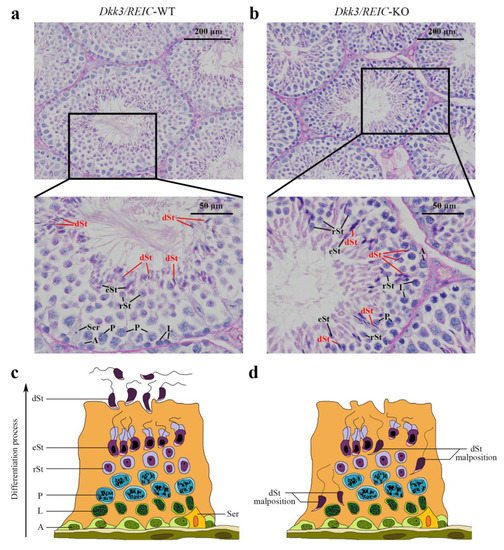
Figure 1. PAS staining of the testicular sections. (a) In the Dkk3/REIC-WT testicular section (stage VIII–IX of spermatogenesis), the distribution of germ cells was well arranged, and the tadpole-like differentiated sperm were uniformly located around the seminiferous tubule lumen waiting to be released. (b) In the Dkk3/REIC-KO testicular section (stage VIII–IX of spermatogenesis), however, the germ cell distribution was much disordered, few mature spermatids were in or surrounding the central lumen, while the differentiated sperm (marked by in red) irregularly wandered adjacent to the basement membrane. (c) The schematic drawing of typical spermatogenesis in the Dkk3/REIC-WT seminiferous epithelium. (d) The schematic drawing of pathological spermatogenesis in the Dkk3/REIC-KO seminiferous epithelium. Abbreviation: A = spermatogonia, Ser = sertoli cell, L = spermatocytes in leptotene phase, P = spermatocytes in pachytene phase, rSt = round spermatids, eSt = elongated spermatids and dSt = differentiated spermatids.
3. Expression and Distribution of Dkk3/REIC in Testes and Sperm of Male Mouse
To explore the transcript expression of Dkk3/REIC in mouse testes, we used the published C57BL/6 testicular scRNA-seq transcriptome in GEO database (GSE112393) and assessed the cell-specific expression of Dkk3/REIC. Clustering analysis identified 11 types of testicular cells (Figure 2a), Dkk3/REIC transcripts existed in most of the cell types except for macrophages and endothelial cells (Figure 2b,c), and this was predominately expressed in spermatogonia cells (Figure 2d). Then, we used testicular sections and sperm smear obtained from KO and WT mice and detected Dkk3/REIC expression by immunofluorescence (IF) staining.
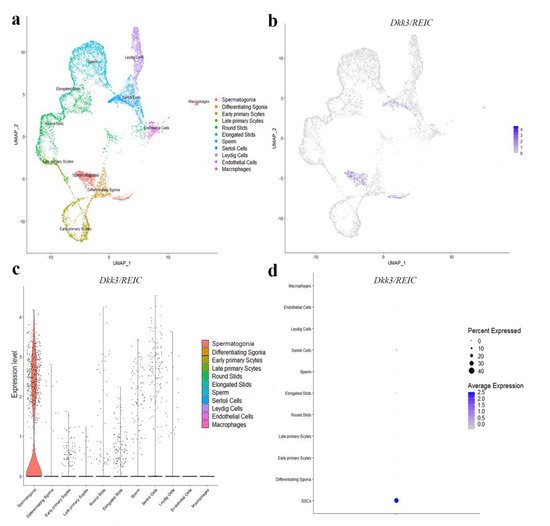
Figure 2. Distribution of Dkk3/REIC among testicular cell clusters in public scRNA-seq transcriptome atlas. (a) Analysis revealed 11 clusters that were matched to 11 testicular cell types. (b) The FeaturePlot function, (c) VlnPlot function and (d) DotPlot function of Seurat packages demonstrated the distribution of Dkk3/REIC among these different cell types. The results indicated that Dkk3/REIC had the highest level in spermatogonia.
As shown in Figure 3a, DKK3/REIC in the testicular section was mainly expressed in spermatogonia. This was compliant with our results from the published scRNA-seq data. Figure 3b shows the distribution of DKK3/REIC in mouse sperm, which was extensively expressed in both the head and tail, indicating that it may be a functional protein for spermatic activity.
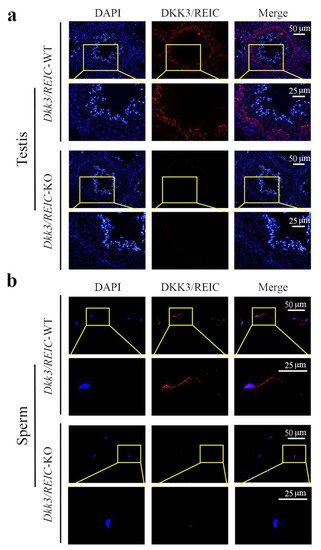
Figure 3. Expression and localization of DKK3/REIC protein in mouse seminiferous tubules and sperm. The expression and distribution of DKK3/REIC protein in (a) testes and (b) sperm of Dkk3/REIC-WT and Dkk3/REIC-KO mice.
4. Sperm Motility Is Impaired in Dkk3/REIC-KO Male Mice While the Sperm Total Count and Vitality Rate Show No Significant Difference
Although the average epididymal sperm count (per epididymis) of Dkk3/REIC-KO mice was nearly 1 million less than their wild-type counterparts (9.30 ± 0.69 vs. 8.27 ± 0.87, ×106), the difference was not statistically significant (p > 0.05). Sperm vitality rates between the WT and KO mice were also non-significant (72.83 ± 1.55% vs. 72.50 ± 0.71%, p > 0.05). Irregular findings were noticed in the motility status of Dkk3/REIC-KO mice sperm. Compared with the wild-types, KO mice showed a significant lower proportion of motile sperm (44.09 ± 8.12% vs. 23.26 ± 10.02%, p < 0.01).
5. Ultrastructural Examination Reveals Defects in Flagellum Fibrous Sheath of Dkk3/REIC-KO Mouse Sperm
The normal sperm structure under TEM is depicted in Figure 4a. Sperm flagellum can be routinely divided into three sub-parts, which are the mid-piece, principal piece and end piece [13]. The cross sections of different sub-parts contain different structures. For Dkk3/REIC-KO sperm, irregular changes were found in the fibrous sheath, a major component of the principal piece. Compared with their wild-type counterparts, which showed an intact fibrous sheath, Dkk3/REIC-KO sperm had multiple defects and fractures in the fibrous sheath of the flagellum (Figure 4b). Such abnormalities could disrupt the structural continuity of the sperm flagellum, which might be linked with the sperm motility impairment in Dkk3/REIC-KO mice.
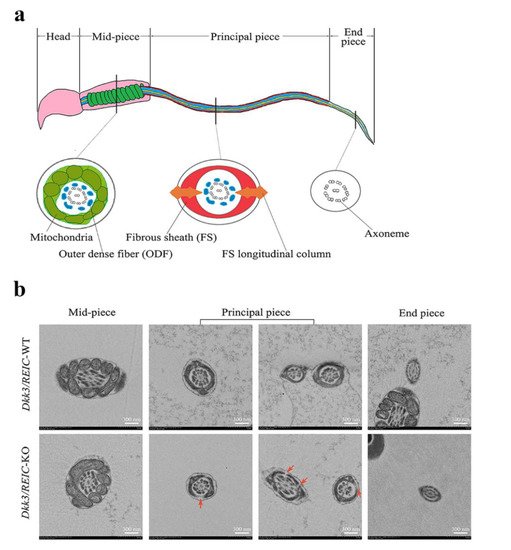
Figure 4. Transmission electron microscope observation of Dkk3/REIC-WT and Dkk3/REIC-KO sperm. (a) Schematic drawing of the mouse sperm ultrastructure. The sperm flagellum can be divided into three parts, which are the mid-piece, principal piece, and end piece. Their cross section views are different from each other, and the fibrous sheath structure mainly exists in the principal piece. (b) The TEM images of Dkk3/REIC-WT and Dkk3/REIC-KO sperm. No obvious difference was found between the mid-piece and end piece; hwoever, the principal piece of Dkk3/REIC-KO sperm showed irregular fractures in the fibrous sheath (arrows).
6. RNA Sequencing of Testicular Transcriptome from Dkk3/REIC-WT and Dkk3/REIC-KO Mice and GO Enrichment Analysis
The RNA-seq of testes transcriptome revealed 36,060 DEGs. The DEGs with FDR value less than 0.05 and a fold change larger than 2 are marked by colors in the volcano plot (Figure 5a). The relative expressions of top 50 DEGs between Dkk3/REIC-WT and Dkk3/REIC-KO testes are shown in a heatmap (Figure 5b). Then, we selected the top 200 DEGs as candidate genes and conducted GO (Gene Ontology) enrichment analysis. As shown in Figure 5c, for the GO category of Biological Process (BP), the candidate genes were predominately enriched in the cellular response to cAMP.
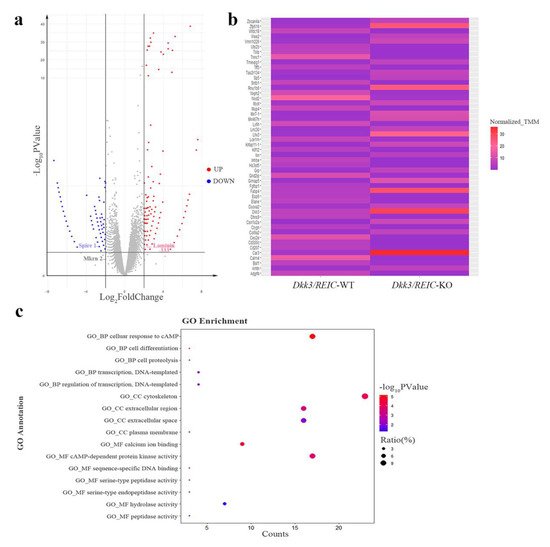
Figure 5. Differentially expressed genes and enrichment results of Dkk3/REIC-WT and Dkk3/REIC-KO mice. (a) Volcano plot of differentially expressed genes (DEGs). Genes with significant difference (FDR < 0.05 and fold change ≥ 2) are marked in colors. Red and blue, respectively, indicate the up-regulated and down-regulated genes in Dkk3/REIC-KO mice. DEGs of Laminin-333, Spire 1 and Mkrn 2 are marked by labels. (b) The relative expressions of the top 50 DEGs among Dkk3/REIC-WT and Dkk3/REIC-KO mice testes. (c) The top 200 DEGs analyzed by Gene Ontology (GO) enrichment.
In the Cellular Component (CC) category, candidates were significantly concentrated in the cytoskeleton. For the Molecular Function (MF) category, calcium ion binding and cAMP-dependent protein kinase activity were the most significantly enriched GO terms. Collectively, the GO enrichment results indicated that the functions of the selected DEGs were mainly focused on cAMP signaling, calcium ion binding and cytoskeleton function, revealing that the absence of Dkk3/REIC may induce changes in the above activities and consequently lead to reproductive impairment.
This entry is adapted from the peer-reviewed paper 10.3390/genes13020285
This entry is offline, you can click here to edit this entry!
