Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Deep learning techniques, such as convolutional neural networks (CNNs), generative adversarial networks (GANs), and graph neural networks (GNNs) have, over the past decade, changed the accuracy of prediction in many diverse fields. In recent years, the application of deep learning techniques in computer vision tasks in pathology has demonstrated extraordinary potential in assisting clinicians, automating diagnoses, and reducing costs for patients.
- deep learning
- machine learning
- histopathology
- computational pathology
1. Classification and Feature Prediction
With the success of CNN models in various real-world computer-vision classification tasks, researchers and scientists have also trained and tested these models in case scenarios in biomedical fields, including pathology. These studies may involve training an existing CNN architecture from scratch. However, it requires more data, and the data augmentation techniques may not always be suitable for biomedical images. Alternatively, transfer learning techniques, which freeze most of the parameters from a model often pre-trained on ImageNet, have more advantages in terms of the data size requirements. For example, an InceptionV3-based ImageNet pre-trained CNN model can achieve a high level of accuracy in determining skin lesion malignancy and the possibility of melanoma [1][2].
In clinical settings, pathologists typically examine histopathology slides under microscopes to provide diagnosis or other clinical information. Due to the development of digital pathology equipment, digitizing histopathology slides is cheaper and more accessible. As a result, more and more deidentified digital histopathology slide images have become available in many databases. These images, often with extremely large dimensions, are saved in special image file formats (e.g., .svs or .scn), which is a tuple of the same image with different resolutions [3]. Thus, in order to fit these digital histopathology images into CNN architectures, people usually develop their own customized pipelines with commonly used techniques, such as tiling the whole slide images (WSI) or sampling regions of interest (ROIs) (Figure 1) [4]. In the past few years, classification CNN models trained on histopathology images have shown phenomenally high performance and promising clinical potential in predicting both morphological features and molecular features. The visualization techniques also reveal results that often match pathologists’ expectations and many models are generalizable to independent real-world clinical images. For example, Inception and InceptionResNet architectural models demonstrate high accuracy and other statistical metrics in predicting subtypes and key biomarker mutations, such as STK11 and EGFR, in non-small-cell lung cancer histopathology slides [5][6][7]. With the integration of other critical clinical variables and images, immune response, G-CIMP, and telomere length can be predicted in glioblastoma patients [8]. BRAF mutation, a well-known biomarker in malignant melanoma, can also be accurately predicted with a CNN-based model [9]. Other molecular and genomic features, such as microsatellite instability (MSI), can be predicted from histopathology slides with a reasonable accuracy as well [10]. The critical gene expression level could also be inferred by applying these CNN classification models to WSI [11]. Some contemporary models also show successful classification results in the histopathology images of multiple tissue types [12][13]. These successful cases indicate that CNNs represent a suitable approach to study the correlation between molecular features and morphological features in histopathology slides, some of which may be undetectable or often ignored by human pathologists.
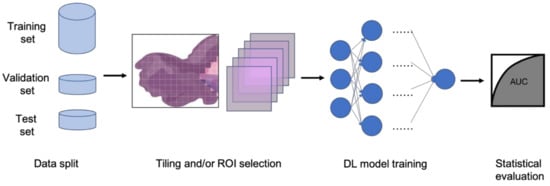
Figure 1. Typical classification model pipeline for histopathology images.
However, histopathology images are quite different from the images in the ImageNet because of their extremely large sizes, higher resolution, and sparser useful feature distributions [14][3][15]. Deep learning architectures that could take advantage of these characteristics are very likely to achieve better results, unveiling more interesting hidden features in histopathology image classification tasks. For instance, a multi-resolution CNN model, which takes advantage of the data structure of .svs and .scn image files, achieves higher performance in classifying endometrial cancer molecular features than its single resolution counterparts [16]. Weakly supervised techniques, such as multiple instance learning, also demonstrate decent performance in classification tasks of histopathology images, and have gained popularity in recent years [15][17][18]. The innovative idea of bringing GNN models into solving histopathology classification problems develops greater capacity in understanding the subtle relationships between features of different tissue structures and at different locations on giant digital histopathology slides [19][20].
2. Segmentation
In addition to classification tasks, CNN models are also capable of segmenting cells or tissue in histopathology slides [21][3]. The segmented cells or tissue could then be used to train classification models for different prediction tasks, including the recurrence of non-small-cell lung cancer [22] and endometrial tissue types [23]. A popular segmentation CNN architecture used in the biomedical field is U-net, which has a similar structure to an autoencoder [24]. A 3D version of U-net, which has 3D convolutional layers instead of 2D convolutional layers, is capable of segmenting volumetric images [25]. Modified U-net architectures, such as USE-net [26], Focus U-net [27], and U-net with attention gate [28], have achieved even better performance in various biomedical image segmentation tasks than vanilla U-net. Other autoencoder-based methods have also achieved promising results in segmentation tasks of histopathology images, such as highlighting tumor regions in liver cancer WSI [29]. Well-trained style transfer models are also viable options for segmentation tasks [30]. With the introduction of GAN, using conditional-GAN or cycle-GAN models and in combination with CNN models for segmentation problems is also shown to be viable, with less stringent training data requirements [31][32][33]. Unlike most classification models, the segmentation models can be more adaptive to different types of tissues due to the similarities of the stained features and textures of the histopathology slides [34]. Additionally, the evaluation metrics of these classification models can be drastically different from those of the classification models. The segmentation labels are also usually images; therefore, it is not easy to determine a binary prediction or even a prediction score at the per-image level. Hence, typical statistical metrics, such as AUROC or precision and recall, are often not capable of fairly evaluating segmentation tasks. Pixel-level metrics, such as intersection over the union (IoU), also pose weaknesses because it cannot objectively give relative importance to pixels of different regions. Object-level metrics can be an optimal alternative, but the requirement of identifying all objects on the label images prohibits its adoption in real-world model evaluation. Therefore, researchers often use customized evaluation metrics with a combination of customized pixel weights, dice loss, and IoU with specific thresholds [35][36].
This entry is adapted from the peer-reviewed paper 10.3390/biomedinformatics2010010
References
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118.
- Puri, M.; Hoover, S.B.; Hewitt, S.M.; Wei, B.-R.; Adissu, H.A.; Halsey, C.H.C.; Beck, J.; Bradley, C.; Cramer, S.D.; Durham, A.C.; et al. Automated Computational Detection, Quantitation, and Mapping of Mitosis in Whole-Slide Images for Clinically Actionable Surgical Pathology Decision Support. J. Pathol. Inform. 2019, 10, 4.
- Cooper, L.A.; Demicco, E.G.; Saltz, J.H.; Powell, R.T.; Rao, A.; Lazar, A.J. PanCancer insights from The Cancer Genome Atlas: The pathologist’s perspective. J. Pathol. 2018, 244, 512–524.
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979.
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567.
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225.e35.
- Hong, R.; Liu, W.; Fenyö, D. Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning. BioMedInformatics 2021, 2, 6.
- Wang, L.B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e20.
- Kim, R.H.; Nomikou, S.; Coudray, N.; Jour, G.; Dawood, Z.; Hong, R.; Esteva, E.; Sakellaropoulos, T.; Donnelly, D.; Moran, U.; et al. Deep learning and pathomics analyses reveal cell nuclei as important features for mutation prediction of BRAF-mutated melanomas. J. Investig. Dermatol. 2021, in press.
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056.
- Schmauch, B.; Romagnoni, A.; Pronier, E.; Saillard, C.; Maillé, P.; Calderaro, J.; Kamoun, A.; Sefta, M.; Toldo, S.; Zaslavskiy, M.; et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020, 11, 3877.
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 2020, 1, 789–799.
- Fu, Y.; Jung, A.W.; Torne, R.V.; Gonzalez, S.; Vöhringer, H.; Shmatko, A.; Yates, L.R.; Jimenez-Linan, M.; Moore, L.; Gerstung, M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 2020, 1, 800–810.
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42.
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309.
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400.
- Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Zhao, M.; Shady, M.; Lipkova, J.; Mahmood, F. AI-based pathology predicts origins for cancers of unknown primary. Nature 2021, 594, 106–110.
- Cinbis, R.G.; Verbeek, J.; Schmid, C. Weakly Supervised Object Localization with Multi-Fold Multiple Instance Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 189–203.
- Anand, D.; Gadiya, S.; Sethi, A. Histographs: Graphs in histopathology. arXiv 2020, arXiv:1908.05020.
- Gao, Z.; Shi, J.; Wang, J. GQ-GCN: Group Quadratic Graph Convolutional Network for Classification of Histopathological Images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; LNCS 2021. Volume 12908.
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175.
- Wang, X.; Velcheti, V.; Vaidya, P.; Bera, K.; Madabhushi, A.; Khunger, A.; Patil, P.; Choi, H. RaPtomics: Integrating Radiomic and Pathomic Features for Predicting Recurrence in Early Stage Lung Cancer. In Medical Imaging 2018: Digital Pathology, Proceedings of the; Gurcan, M.N., Tomaszewski, J.E., Eds.; SPIE: Bellingham, WA, USA, 2018; Volume 10581, p. 21.
- Sun, H.; Zeng, X.; Xu, T.; Peng, G.; Ma, Y. Computer-Aided Diagnosis in Histopathological Images of the Endometrium Using a Convolutional Neural Network and Attention Mechanisms. IEEE J. Biomed. Heal. Inform. 2020, 24, 1664–1676.
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2015, 9351, 234–241.
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation; Springer: Cham, Switzerland, 2016; pp. 424–432.
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.S.; Ferretti, C.; Besozzi, D.; et al. USE-Net: Incorporating Squeeze-and-Excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets. Neurocomputing 2019, 365, 31–43.
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Focus U-Net: A novel dual attention-gated CNN for polyp segmentation during colonoscopy. Comput. Biol. Med. 2021, 137, 104815.
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207.
- Roy, M.; Kong, J.; Kashyap, S.; Pastore, V.P.; Wang, F.; Wong, K.C.L.; Mukherjee, V. Convolutional autoencoder based model HistoCAE for segmentation of viable tumor regions in liver whole-slide images. Sci. Rep. 2021, 11, 139.
- Hollandi, R.; Szkalisity, A.; Toth, T.; Tasnadi, E.; Molnar, C.; Mathe, B.; Grexa, I.; Molnar, J.; Balind, A.; Gorbe, M.; et al. A deep learning framework for nucleus segmentation using image style transfer. bioRxiv 2019, 580605.
- Yi, X.; Walia, E.; Babyn, P. Generative Adversarial Network in Medical Imaging: A Review. arXiv 2018, arXiv:1809.07294.
- Fu, C.; Lee, S.; Joon Ho, D.; Han, S.; Salama, P.; Dunn, K.W.; Delp, E.J. Three Dimensional Fluorescence Microscopy Image Synthesis and Segmentation. arXiv 2018, arXiv:1801.07198.
- Ho, D.J.; Fu, C.; Salama, P.; Dunn, K.W.; Delp, E.J. Nuclei Segmentation of Fluorescence Microscopy Images Using Three Dimensional Convolutional Neural Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017.
- Greenwald, N.F.; Miller, G.; Moen, E.; Kong, A.; Kagel, A.; Camacho Fullaway, C.; Mcintosh, B.J.; Leow, K.; Schwartz, M.S.; Dougherty, T.; et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. bioRxiv 2021.
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Csaba, M.; McQuin, C.; Singh, S.; Theis, F.; et al. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. bioRxiv 2019, 335216.
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations; Springer: Cham, Switzerland, 2017; pp. 240–248.
This entry is offline, you can click here to edit this entry!
