Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
|
Veterinary Sciences
Torque teno sus virus (TTSuV) belongs to the Anelloviridae family. TTSuV is a non-enveloped circular ssDNA virus which frequently infects swine and has been associated with hepatic, respiratory, and autoimmune disorders. TTSuV pathogenic role is still uncertain, and clear data in the literature on virus reservoirs are lacking.
- swine-related virus
- wild ungulates
- reservoir
1. Introduction
Torque teno viruses (TTVs) belong to the Anelloviridae family and are widespread worldwide in humans and animals [1][2]. TTVs were isolated for the first time in humans in 1997 in a patient with post-transfusional hepatitis of unknown etiology [3]. Human infection has been associated with respiratory diseases, acute enteritis, viral hepatitis, and autoimmune rheumatic diseases [4][5][6][7]. Their pathological mechanisms are still unclear, but recent studies suggested an interaction with the host immune system. Moreover, in the case of co-infection with other viruses an increased disease severity was reported [8].
TTVs are ubiquitous viruses and have been often detected in mammalian species, including dogs, cats, swine, cattle, sheep, wild boars, hares, and non-human primates [9][10][11][12][13][14]. TTVs are genetically distinct and are classified in a species-specific manner, although the genomes of TTVs detected from several animal species, including humans, show a similar organization [15].
In swine, Torque teno sus virus (TTSuV) has a genome organization similar to human TTV (huTTV) and comprises two genera: Iotatorquevirus, which includes TTSuV1a, and Kappatorquevirus, which includes two species (TTSuVk2a and TTSuVk2b [2][16][17][18][19]. TTSuV1a and TTSuVk2a are the most studied and well-characterized genogroups and appear to act as primary pathogens in swine, causing mild to moderate respiratory, hepatic, and nephritic lesions [15][20][21]. Recently, TTSuV co-infection with other swine-related viruses was studied, and it was demonstrated that the association of TTSuV with Porcine circovirus type 2 can enhance disease severity [22].
TTSuVs are mainly transmitted by the fecal–route, and they are frequently detected in fecal excretions as well as nasal excretions, sera, and several organs including the liver of infected pigs [23]. The virus transmission may also occur by a vertical route, as fetuses infected with TTSuV have been found at different stages of pregnancy [24][25].
TTSuV is widespread in Europe in both swine and wild boars, as reported by different studies: TTSuV was found in Romania and in Germany, while a high sero-prevalence (84%) was found among wild boars in Spain [12][26][27]. In Italy, previous studies reported a prevalence of 83.2% in sera of pigs at different ages, as well as a prevalence of 58.3% in fresh pork liver sausages [28][29][30]. Moreover, a recent study investigated the role of wild animals as TTSuV reservoirs in Northern Italy. TTSuV prevalence in wild boars was as high as 4.5%, while in wild ruminants a prevalence of 9.4% was reported [31], highlighting the need for monitoring studies in wild populations to improve the knowledge of emerging viruses and to better evaluate their likely zoonotic potential. In fact, wild animals could be putative reservoirs that may contribute, to varying degrees, to the spread and amplification of viruses, possibly including those initially originated on farms, to other farms with eventual transmission to humans. The public health significance of TTSuVs as potentially zoonotic swine viruses must be evaluated, considering that modern human lifestyle, which is characterized by growing populations, high density rates, and global movements, leads to the increase in new risks for human and animal health.
2. TTSuV Prevalence in Wild Ungulates and Associated Risk Factors
A total of 528 wild ungulate liver samples were analyzed (n = 400 from wild boars, and n = 128 from wild ruminants), and the overall prevalence was calculated (Table 1).
Table 1. TTSuV prevalence and 95% confidence intervals (95% CI) are reported. Results are divided by species (wild ruminants include red deer, roe deer, and chamois), risk factors evaluated (animal gender and age class), sampling area, and hunting season.
| Wild Ungulates (n = Samples) | TTSuV Prevalence (95% CI, n = Positive Samples) |
|---|---|
| Overall (n = 528) | 5.7% (3.9–8.0%, n = 30) |
| Species | |
| Wild ruminants (n = 128) | 9.4% (4.9–15.8%, n = 12) |
| Wild boars (n = 400) | 4.5% (2.7–7.0%, n = 18) |
| Gender | |
| Female (n = 259) | 4.2% (2.1–7.5%, n = 11) |
| Male (n = 269) | 7.1% (4.3–10.8%, n = 19) |
| Age class 1 | |
| 0 (n = 96) | 8.3% (3.7–15.8%, n = 8) |
| 1 (n = 196) | 5.1% (2.5–9.2%, n = 10) |
| 2 (n = 236) | 5.1% (2.6–8.7%, n = 12) |
| Sampling area | |
| Parma (n = 175) | 2.3% (0.6–5.7%, n = 4) |
| Sondrio (n = 353) | 7.4% (4.9–10.6%, n = 26) |
| Hunting season | |
| 2016–2017 (n = 122) | 13.9% (8.3–21.4%, n = 17) |
| 2017–2018 (n = 32) | 9.4% (2.0–25.0%, n = 3) |
| 2018–2019 (n = 225) | 2.7% (1.0–5.7%, n = 6) |
| 2019–2020 (n = 149) | 2.7% (0.7–6.7%, n = 4) |
1 Class 0: young; class 1: sub-adult; class 2: adult.
Overall TTSuV prevalence was 5.7% (30/528, 95% CI 3.9–8.0%). A higher prevalence (p-value < 0.05) was found in wild ruminants (9.4%, 95% CI 4.9–15.8%) compared to wild boars (4.5%, 95% CI 2.7–7.0%). No risk factor was associated between TTSuV presence and either gender or age class of wild animals (p-value > 0.05). Considering sampling areas, a higher prevalence (p-value < 0.05) was found in wild ungulates in Sondrio Province (7.4%, 95% CI 4.9–10.6%) compared to the Parma area (2.3%, 95% CI 0.6–5.7%); TTSuV prevalence in wild boars (data not shown) was also higher in the mountains than in the flatland (14/225, 6.2%, 95% CI 3.7–10.2 in Sondrio Province, and 4/175, 2.3%, 95% CI 0.6–5.7% in the Parma area). Finally, the highest prevalence was observed in the 2016–2017 hunting season (13.9%, 95% CI 8.3–21.4%), followed by a decrease throughout the 2019–2020 hunting season (2.7%, 95% CI 0.7–6.7%), (p-value < 0.05).
3. TTSuV1a and TTSuVk2a Prevalence
TTSuV1a and TTSuVk2a prevalence was investigated in wild boars and wild ruminants according to gender, age class, hunting season, and sampling area.
The overall prevalence in wild ungulates was 2.1% (11/528, CI95% 1.2–3.7%) for TTSuV1a and 4.2% (22/528, CI95% 2.8–6.2%) for TTSuVk2a (data not shown).
Results for TTSuV1a and TTSuVk2a detection in wild boars are shown in Table 2.
Table 2. TTSuV1a and TTSuVk2a prevalence and 95% confidence intervals (95% CI) according to gender, age class, hunting season, and sampling area.
| Wild Boars (n = Samples) | TTSuV1a Prevalence (95%CI, n = Positive Samples) |
TTSuVk2a Prevalence (95%CI, n = Positive Samples) |
|---|---|---|
| Overall (n = 400) | 2.7% (1.4–4.9%, n = 11) | 2.5% (1.2–4.5%, n = 10) |
| Gender | ||
| Males (n = 216) | 3.2% (1.3–6.6%, n = 7) | 3.7% (1.6–7.2%, n = 8) |
| Females (n = 184) | 2.2% (0.6–5.5%, n = 4) | 1.1% (0.1–3.9%, n = 2) |
| Age class 1 | ||
| 0 (n = 60) | 1.7% (0–8.9%, n = 1) | 5.0% (1.0–13.9%, n = 3) |
| 1 (n = 170) | 2.9% (1.0–6.7%, n = 5) | 2.9% (1.0–6.7%, n = 5) |
| 2 (n = 170) | 2.9% (1.0–6.7%, n = 5) | 1.2% (0.1–4.2%, n = 2) |
| Hunting season | ||
| 2018–2019 (n = 169) | 5.3% (2.8–9.8%, n = 9) | 4.1% (2.0–8.3%, n = 7) |
| 2019–2020 (n = 231) | 0.9% (0.2–3.1%, n = 2) | 1.3% (0.4–3.7%, n = 3) |
| Sampling area | ||
| Sondrio (n = 225) | 3.6% (1.5–6.9%, n = 8) | 3.6% (1. 5–6.9%, n = 8) |
| Parma (n = 175) | 1.7% (0.3–4.9%, n = 3) | 1.1% (0.1–4.1%, n = 2) |
1 Class 0: young; class 1: sub-adult; class 2: adult.
In wild boars, TTSuV1a was detected in 11/400 samples (2.7%, CI95% 1.4–4.9%), while TTSuVk2a was detected in 10/400 samples (2.5%, CI95% 1.2–4.5%). In three wild boar samples (0.75%, CI95% 0.3–2.2%) TTSuV1a and TTSuVk2a co-infection was observed; two samples were found in Sondrio Province and one in the Parma area (data not shown).
Considering wild boar gender, TTSuV1a and TTSuVk2a prevalence in males was 3.2% (CI95% 1.3–6.6%) and 3.7% (CI95% 1.6–7.2%), respectively. In females, TTSuV1a prevalence was 2.2% (CI95% 0.6–5.5%), while TTSuVk2a prevalence was 1.1% (CI95% 0.1–3.9%). No positive hepatic sample was found in pregnant animals. Moreover, TTSuV1a was detected in 1.7% (CI95% 0–8.9%) of class 0 animals and in 2.9% (CI95% 1.0–6.7%) of both class 1 and 2 animals. TTSuVk2a was detected with 5.0% prevalence (CI95% 1.0–13.9%) in class 0, 2.9% (CI95% 1.0–6.7%) in class 1, and 1.2% (CI95% 0.1–4.2%) in class 2.
In the 2018–2019 hunting season, a prevalence of 5.3% (CI95% 2.8–9.8%) and 4.1% (CI95% 2.0–8.3%) was observed for TTSuV1a and TTSuVk2a, respectively, while in the 2019––2020 hunting season prevalence decreased to 0.9% (CI95% 0.2–3.1%) for TTSuV1a, and 1.3% (CI95% 0.4–3.7%) for TTSuVk2a (Table 2).
In wild boars from Sondrio Province, both TTSuV1a and TTSuVk2a prevalence was 3.6% (CI95% 1.5-6.9%), while in the Parma area TTSuV1a was detected in 1.7% (CI95% 0.3–4.9%) samples, and TTSuVk2a in 1.1% (CI95% 0.1–4.1%) samples (Table 2).
In wild ruminants (red deer, roe deer, and chamois) from Sondrio Province, TTSuV1a was not detected, while TTSuVk2a prevalence was 9.4% (CI95% 4.9–15.8%) (Table 3).
Table 3. TTSuVk2a prevalence and 95% confidence intervals (95% CI) in wild ruminants according to gender, age class, and hunting season.
| Wild Ruminants (n = Samples) |
TTSuVk2a Prevalence (95% CI, n = Positive Samples) |
|---|---|
| Overall (n = 128) | 9.4% (4.9–15.8%, n = 12) |
| Gender | |
| Males (n = 53) | 11.3% (4.3–23.0%, n = 6) |
| Females (n = 75) | 8.0% (3.0–16.6%, n = 6) |
| Age class 1 | |
| 0 (n = 36) | 11.1% (3.1–26.1%, n = 4) |
| 1 (n = 26) | 7.7% (0.9–25.1%, n = 2) |
| 2 (n = 66) | 9.1% (3.4–18.7%, n = 6) |
| Hunting season | |
| 2016–2017 (n = 58) | 13.8% (6.1–25.4%, n = 8) |
| 2017–2018 (n = 32) | 9.4% (2.0–25.0%, n = 3) |
| 2018–2019 (n = 38) | 2.6% (0.1–13.8%, n = 1) |
1 Class 0: young; class 1: sub-adult; class 2: adult.
TTSuVk2a showed a prevalence of 11.3% (CI95% 4.3–23.0%) in males and 8.0% (CI95% 3.0–16.6%) in females. TTSuVk2a prevalence in class 0 was 11.1% (Cl95% 3.1–26.1%); in class 1 it was 7.7% (Cl95% 0.9–25.1%), and in class 2 it was 9.1% (Cl95% 3.4–18.7%). From 2016–1017 to 2018–2019 the prevalence decreased from 13.8% (Cl95% 6.1–25.4%) to 2.6% (Cl95% 0.1–13.8%).
4. Phylogenetic Tree
The phylogenetic analysis of TTSuV was performed on sequences obtained from 10 wild boars and 1 red deer liver sample, confirming the presence of both genogroups 1 and 2 in wild animals (Figure 1). In particular, TTSuV1a and TTSuVk2a were detected in 3 and 7 wild boar liver samples, respectively. In red deer, only one TTSuVk2a strain was detected. All sequences were obtained from animals infected with one viral genogroup, except for sample B1_16 (wild boar) with a TTSuV1a/TTSuVk2a co-infection.
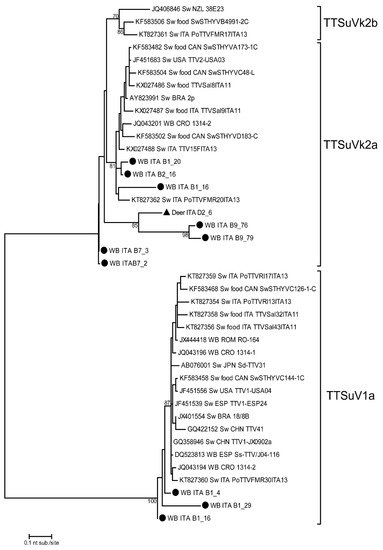
Figure 1. Phylogenetic tree of TTSuV 5′- end of the UTR (nt 138–453) nucleotide sequences. TTSuV strains of this study are indicated with a black circle for wild boars and a black triangle for red deer. Reference sequences are also considered in the dendrogram. The maximum likelihood phylogenetic tree was constructed with 1000 bootstrap repetitions. Bootstrap values under 70% are not shown. WB = wild boar; Sw = swine.
The phylogenetic tree in Figure 1 reveals a high heterogeneity among the obtained nucleotide sequences. The wild boar strains belonging to TTSuV1a (B1_4, B1_16, and B1_29) had the same geographical and temporal origin (Sondrio Province in the 2018–2019 hunting season). These sequences shared an intra-group nucleotide identity (nt. id.) ranging between 86.2% and 96.7% and revealed the highest nt. id. with two TTSuV1a strains previously detected in Croatia (JQ043194) and Italy (KT827360) in wild boar and swine liver samples, respectively, and are related also to food products containing raw swine liver. The phylogenetic tree also shows a high nucleotide sequence heterogeneity when considering the TTSuVk2a clade. In fact, TTSuV strains detected in this study in wild boar and red deer were grouped in three distinct clades related to strains from several animal hosts. The first clade included three strains (B1_16, B1_20, and B2_16) detected in wild boars (clade nt. id. ranging between 88.3% and 97.5%) grouped with strains detected in domestic pigs in Italy and in wild boars in Croatia. The three strains from this study were all collected in Sondrio Province in the 2018–2019 hunting season. The second (B9_76, B9_79, and D2_6) and third clades (B7_2 and B7_3) included three (two from wild boars collected in the Parma area in 2019–2020, and one from red deer collected in Sondrio Province in 2017–2018) and two (from wild boars collected in the Parma area in 2019–2020) sequences showing between 75.6% and 84.5% nt. id. with other TTV reference strains.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10020242
References
- Gallian, P.; Biagini, P.; Zhong, S.; Touinssi, M.; Yeo, W.; Cantaloube, J.F.; Attoui, H.; de Micco, P.; Johnson, P.J.; de Lamballerie, X. TT Virus: A Study of Molecular Epidemiology and Transmission of Genotypes 1, 2 and 3. J. Clin. Virol. 2000, 17, 43–49.
- International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/ssdna-viruses-2011/w/ssdna_viruses/139/anelloviridae (accessed on 4 December 2021).
- Nishizawa, T.; Okamoto, H.; Konishi, K.; Yoshizawa, H.; Miyakawa, Y.; Mayumi, M. A Novel DNA Virus (TTV) Associated with Elevated Transaminase Levels in Posttransfusion Hepatitis of Unknown Etiology. Biochem. Biophys. Res. Commun. 1997, 241, 92–97.
- Prasetyo, A.A.; Desyardi, M.N.; Tanamas, J.; Suradi; Reviono; Harsini; Kageyama, S.; Chikumi, H.; Shimizu, E. Respiratory Viruses and Torque Teno Virus in Adults with Acute Respiratory Infections. Intervirology 2015, 58, 57–68.
- Brassard, J.; Gagné, M.-J.; Leblanc, D.; Poitras, É.; Houde, A.; Boras, V.F.; Inglis, G.D. Association of Age and Gender with Torque Teno Virus Detection in Stools from Diarrheic and Non-Diarrheic People. J. Clin. Virol. 2015, 72, 55–59.
- Kasirga, E.; Sanlidag, T.; Akcali, S.; Keskin, S.; Aktas, E.; Karakoç, Z.; Helvaci, M.; Sözen, G.; Kuzu, M. Clinical Significance of TT Virus Infection in Children with Chronic Hepatitis B. Pediatrics Int. Off. J. Jpn. Pediatric Society 2005, 47, 300–304.
- Gergely, P.; Perl, A.; Poór, G. Possible Pathogenic Nature of the Recently Discovered TT Virus: Does It Play a Role in Autoimmune Rheumatic Diseases? Autoimmun. Rev. 2006, 6, 5–9.
- Spandole, S.; Cimponeriu, D.; Berca, L.M.; Mihăescu, G. Human Anelloviruses: An Update of Molecular, Epidemiological and Clinical Aspects. Arch. Virol. 2015, 160, 893–908.
- Leary, T.P.; Erker, J.C.; Chalmers, M.L.; Desai, S.M.; Mushahwar, I.K. Improved Detection Systems for TT Virus Reveal High Prevalence in Humans, Non-Human Primates and Farm Animals. J. Gen. Virol. 1999, 80(Pt. 8), 2115–2120.
- Cong, M.; Nichols, B.; Dou, X.; Spelbring, J.E.; Krawczynski, K.; Fields, H.A.; Khudyakov, Y.E. Related TT Viruses in Chimpanzees. Virology 2000, 274, 343–355.
- Okamoto, H.; Takahashi, M.; Nishizawa, T.; Tawara, A.; Fukai, K.; Muramatsu, U.; Naito, Y.; Yoshikawa, A. Genomic Characterization of TT Viruses (TTVs) in Pigs, Cats and Dogs and Their Relatedness with Species-Specific TTVs in Primates and Tupaias. J. Gen. Virol. 2002, 83(Pt. 6), 1291–1297.
- Martínez, L.; Kekarainen, T.; Sibila, M.; Ruiz-Fons, F.; Vidal, D.; Gortázar, C.; Segalés, J. Torque Teno Virus (TTV) Is Highly Prevalent in the European Wild Boar (Sus Scrofa). Vet. Microbiol. 2006, 118, 223–229.
- Brassard, J.; Gagné, M.-J.; Lamoureux, L.; Inglis, G.D.; Leblanc, D.; Houde, A. Molecular Detection of Bovine and Porcine Torque Teno Virus in Plasma and Feces. Vet. Microbiol. 2008, 126, 271–276.
- Águeda-Pinto, A.; Kraberger, S.; Lund, M.C.; Gortázar, C.; McFadden, G.; Varsani, A.; Esteves, P.J. Coinfections of Novel Polyomavirus, Anelloviruses and a Recombinant Strain of Myxoma Virus-MYXV-Tol Identified in Iberian Hares. Viruses 2020, 12, 340.
- Ssemadaali, M.A.; Effertz, K.; Singh, P.; Kolyvushko, O.; Ramamoorthy, S. Identification of Heterologous Torque Teno Viruses in Humans and Swine. Sci. Rep. 2016, 6, 26655.
- Varsani, A.; Opriessnig, T.; Celer, V.; Maggi, F.; Okamoto, H.; Blomström, A.L.; Cadar, D.; Harrach, B.; Biagini, P.; Kraberger, S. Taxonomic update for mammalian anelloviruses (family Anelloviridae). Arch. Virol. 2021, 166, 2943–2953.
- Manzin, A.; Mallus, F.; Macera, L.; Maggi, F.; Blois, S. Global Impact of Torque Teno Virus Infection in Wild and Domesticated Animals. J. Infect. Dev. Ctries. 2015, 9, 562–570.
- Liu, J.; Guo, L.; Zhang, L.; Wei, Y.; Huang, L.; Wu, H.; Liu, C. Three New Emerging Subgroups of Torque Teno Sus Viruses (TTSuVs) and Co-Infection of TTSuVs with Porcine Circovirus Type 2 in China. Virol. J. 2013, 10, 189.
- Cortey, M.; Pileri, E.; Segalés, J.; Kekarainen, T. Globalisation and Global Trade Influence Molecular Viral Population Genetics of Torque Teno Sus Viruses 1 and 2 in Pigs. Vet. Microbiol. 2012, 156, 81–87.
- Krakowka, S.; Ellis, J.A. Evaluation of the Effects of Porcine Genogroup 1 Torque Teno Virus in Gnotobiotic Swine. Am. J. Vet. Res. 2008, 69, 1623–1629.
- Krakowka, S.; Hartunian, C.; Hamberg, A.; Shoup, D.; Rings, M.; Zhang, Y.; Allan, G.; Ellis, J.A. Evaluation of Induction of Porcine Dermatitis and Nephropathy Syndrome in Gnotobiotic Pigs with Negative Results for Porcine Circovirus Type 2. Am. J. Vet. Res. 2008, 69, 1615–1622.
- Ellis, J.A.; Allan, G.; Krakowka, S. Effect of Coinfection with Genogroup 1 Porcine Torque Teno Virus on Porcine Circovirus Type 2-Associated Postweaning Multisystemic Wasting Syndrome in Gnotobiotic Pigs. Am. J. Vet. Res. 2008, 69, 1608–1614.
- Li, G.; Wang, R.; Cai, Y.; Zhang, J.; Zhao, W.; Gao, Q.; Franzo, G.; Su, S. Epidemiology and Evolutionary Analysis of Torque Teno Sus Virus. Vet. Microbiol. 2020, 244, 108668.
- Sibila, M.; Martínez-Guinó, L.; Huerta, E.; Llorens, A.; Mora, M.; Grau-Roma, L.; Kekarainen, T.; Segalés, J. Swine Torque Teno Virus (TTV) Infection and Excretion Dynamics in Conventional Pig Farms. Vet. Microbiol. 2009, 139, 213–218.
- Aramouni, M.; Segalés, J.; Cortey, M.; Kekarainen, T. Age-Related Tissue Distribution of Swine Torque Teno Sus Virus 1 and 2. Vet. Microbiol. 2010, 146, 350–353.
- Gallei, A.; Pesch, S.; Esking, W.S.; Keller, C.; Ohlinger, V.F. Porcine Torque Teno Virus: Determination of Viral Genomic Loads by Genogroup-Specific Multiplex Rt-PCR, Detection of Frequent Multiple Infections with Genogroups 1 or 2, and Establishment of Viral Full-Length Sequences. Vet. Microbiol. 2010, 143, 202–212.
- Cadar, D.; Kiss, T.; Ádám, D.; Cságola, A.; Novosel, D.; Tuboly, T. Phylogeny, Spatio-Temporal Phylodynamics and Evolutionary Scenario of Torque Teno Sus Virus 1 (TTSuV1) and 2 (TTSuV2) in Wild Boars: Fast Dispersal and High Genetic Diversity. Vet. Microbiol. 2013, 166, 200–213.
- Blois, S.; Mallus, F.; Liciardi, M.; Pilo, C.; Camboni, T.; Macera, L.; Maggi, F.; Manzin, A. High Prevalence of Co-Infection with Multiple Torque Teno Sus Virus Species in Italian Pig Herds. PLoS ONE 2014, 9, e113720.
- Martelli, F.; Caprioli, A.; Di Bartolo, I.; Cibin, V.; Pezzotti, G.; Ruggeri, F.M.; Ostanello, F. Detection of Swine Torque Teno Virus in Italian Pig Herds. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 234–238.
- Monini, M.; Vignolo, E.; Ianiro, G.; Ostanello, F.; Ruggeri, F.M.; Di Bartolo, I. Detection of Torque Teno Sus Virus in Pork Bile and Liver Sausages. Food Environ. Virol. 2016, 8, 283–288.
- Francesco Righi; Sara Arnaboldi; Virginia Filipello; Giovanni Ianiro; Ilaria Di Bartolo; Stefania Calò; Silvia Bellini; Tiziana Trogu; Davide Lelli; Alessandro Bianchi; et al. Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy. Microorganisms 2022, 10, 242, 10.3390/microorganisms10020242.
This entry is offline, you can click here to edit this entry!
