Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Sulfur is a growth-limiting and secondary macronutrient as well as an indispensable component for several cellular components of crop plants.
- sulfur
- sulfate transporters
- seed storage proteins
1. Introduction
Sulfur is an essential macronutrient; according to its chemical nature it plays important roles in various cellular metabolic processes [1]. As an organic thiol it is incorporated in essential amino acids such as cysteine, methionine and different co-enzymes (biotin, coenzyme A, iron-sulfur proteins, ethylene, thiamine pyrophosphate and lipoic acid), and thioredoxins and sulfolipids are often responsible for the structure and biological activity of proteins [2][3]. It has been noted that a deficiency of sulfur in soil is a problematic issue which leads to reduction in yield and quality of seeds. Day by day the concentration of sulfur in soil is decreasing due to the increasing proportions of high analysis and sulfur-free fertilizers and the decreasing use of traditional organic manures in agricultural land. Sulfur is taken up by a plant’s roots in the form of sulfate from the soil, which is also a transportable form in the plant system [4]. Moreover, Abou Seeda et al. [2] revealed that the primary uptake mechanism of sulfate takes place via roots, and afterwards, it is translocated to various locations within the plant body through xylem. Assimilation of sulfur is occurring in the form of cysteine; it acts as a precursor/donor of reduced sulfur mostly for other several organic sulfur compounds present in plants [5]. In rice plants leaf, a significant increment in carbohydrates (starch) was noted when the plants were grown in sulfur deficient soil [6].
2. Source to Sink Relationship
Sulfur is taken up from the rhizospheric region by plant roots as sulfate and distributed within the tissues in this form. Moreover, Tabe and Droux [7] depicted that the dominant form of sulfur is sulfate and it is translocated via phloem, supplied in pods during the time of lupin (Lupinus albus L.) seed development, then the seed is able to reduce and assimilate sulfate. Within the plant tissue, including the developing phase of seeds, if sulfate is not in a reduced form, then the excess amount of sulfate is stored in the vacuole [8][9]. In developing seeds of M. truncatula, a transcriptomic analysis revealed that within the seed tissues sulfur assimilation takes place by following two distinct pathways (Figure 1). In most of the cases, sulfate enters the embryo and is utilized for the biosynthesis of cysteine (Cys) and gets incorporated into proteins, while in another pathway sulfate enters into the seed coat and endosperm. The latter is preferentially involved in the biosynthesis of defense-related sulfur compounds [10]. This kind of partitioning of sulfur in between the seed compartment implies an active exchange of sulfate.
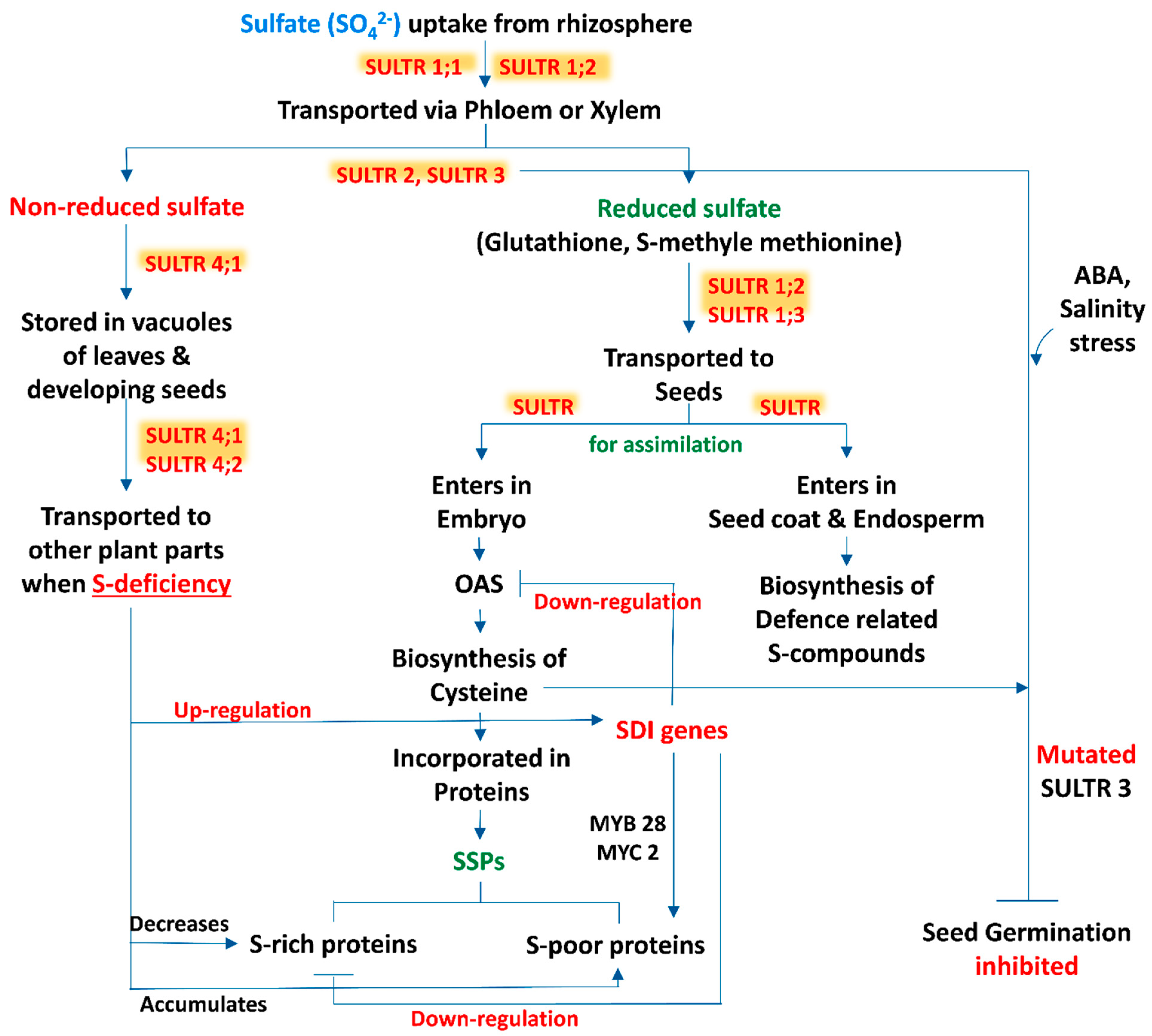
Figure 1. A schematic representation of source-sink relationship of sulfur (S) in plants: an insight in uptake and transportation of sulfate (SO42−) in different plant parts by various transporters (SULTRs) and its assimilation, accumulation of S-containing seed storage proteins (SSPs) and regulation of S-mediated seed germination. (Abbreviations: OAS, O-acetylserine; SDI genes, sulfur deficiency induced genes; MYB28, myeloblastosis28; MYC2, master regulator of cell cycle entry and proliferative metabolism2; ABA, abscisic acid).
3. Sulfate Transporters: Regulate the Sulfur Translocation in Seeds
The beginning of the sulfate uptake mechanism by the root tissue from the surrounding environment and the translocation of sulfate between different cell compartments is facilitated by specific sulfate transporters (SULTR) (Figure 1). The expression levels of these transporter genes in specific organs, cell types and subcellular compartments are regulated by the transcriptional and post transcriptional mechanism which maintains the homeostatic balance between the uptake of sulfate and internal tissue distribution on the basis of sulfate availability and on-demand organic sulfur metabolites biosynthesis [11]. SULTR, a large gene family, is involved in encoding this transporter which consists of 14 members in Arabidopsis and rice (Oryza sativa L.). According to phylogenetic studies this gene family can be categorized into 4 closely related groups (SULTR1 to 4), each containing 12 membrane-spanning domains and a STAS (sulfate transporter and anti-sigma antagonist) domain at the carboxy-terminal end [12], and a fifth member of this group is SULTR5, distinct from the other, lacking that STAS domain [13]. Interestingly, Tomatsu et al. [14] mentioned that the Arabidopsis Sultr5;2 gene was responsible for encoding a high-affinity root molybdate transporter, which raises a valid question about the role of group 5 transporter genes in the sulfate transport mechanism. Furthermore, group 1 and 2 transporters of sulfate are localized in the plasma membrane and are considered as the best categorized groups, being subjected to several studies. Members of group 1 sulfate transporters represent a high-affinity transport system which facilitate the uptake of sulfate by roots (SULTR1;1 and SULTR1;2) or translocate the sulfate from source to sink tissues (SULTR1;3) [15][16][17][18][19]. In addition, the group 2 members consist of low-affinity sulfate transporters whose gene products may be involved in vascular tissues transportation and facilitate the translocation of sulfate throughout the plant [20][13][18]. The literature suggests that group 3 is composed of low-affinity transporters localized at the plasma membrane, showing differential expression patterns in plant tissues and not stimulated by sulphur deficiency [13]. In addition, a role of the SULTR3;5 transporter is noted in the transport of sulfate from root-to-shoot in cooperation with the SULTR2;1 transporter of Arabidopsis [21]. The last group, i.e., group 4 sulfate transporters, has been identified in the vacuolar membrane: a study with SULTR4;1-GFP fusion protein showed that this transporter mainly accumulated in the vacuoles of roots and hypocotyls of young seedlings [21]. Under sulphur sufficient and deficient conditions, the Sultr4;1 transporter gene was expressed in roots and helps to efflux sulfate (SO42-) from the vacuolar lumen to cytoplasm, and it also improved the storage capacity of sulfate in vacuoles [21]. In contrast, gene expression of Sultr4;2 was highly inducible by sulfur during the time of sulfur deficient condition in the same tissue. The double knock-out mutants, i.e., sultr4;1/sultr4;2, contained higher amounts of sulfate as compared to wild-type plants. Comparison between the single and double knock-out mutants sultr4;1/sultr4;2 demonstrated that Sultr4;1 devotes a vital role and Sultr4;2 has a supplementary effect [22]. Whereas the sulfate transport system has been studied extensively in roots, to date, there are very few reports available based on the functions of individual sulfate transporters within seeds. Moreover, vacuoles may play an important role for the purpose of storage and unloading of sulfate within the developing seed, where the members of the SULTR4 transporters would play a key role. In this context, the Arabidopsis Sultr4;1 gene was expressed strongly within the developing seeds, and it was observed that its disruption significantly enhanced the content of seed sulfate, depicting that SULTR4;1 was involved in the efflux mechanism of sulfate from vacuoles to the developing seeds. In addition, a proteomic study of Sultr4;1 mutant seeds revealed the metabolic adjustment for the adaptations in altered sulfate compartmentalization, which indicates a SULTR4;1-mediated sulfate transport system for the establishment of defense mechanisms against oxidative stress during the time of seed development.
For instance, in mature mutant seeds of sultr4;1, on average, the sulfate content was 1.7 times more than that of wild-type plants, whereas the total sulfur content in seeds remained unchanged. In mature seeds, sulfate contributes a significant fraction of the total sulfur content, i.e., 7.7% in wild-type and 13.24% in sultr4;1 mutant seeds, respectively. In respect to mutant sultr4;1 plant phenotypic response, no significant difference was observed in terms of yield parameters, leaf area and/or onset of flowering but a slight reduction in seed weight was noted as compared to wild-type plants. The sulfur ion fluxes into the developing seeds may not be affected in the sultr4;1 mutant, which increases the sulfate contents in seed, but there is no relation for such a drastic perturbation of vegetative growth of this mutant. In mature seeds of the sultr4;1 mutant, a significant increment of sulfate pool was observed, which may be related to a reduced efflux of sulfate from the vacuoles during the development of seeds. In continuation with this, in the reproductive growth phase of Arabidopsis, the Sultr4;1 transporter was preferentially expressed in developing seeds during the time of transition between embryogenesis and the seed filling phase and a relatively higher amount of transcript availability was observed in comparison to Sultr4;2, which was expressed at equal levels throughout the development of seeds. Additionally, the transporter SULTR4;1 also maintains the redox homeostatic balance at the time of seed development if any kind of oxidative stress outbreak takes place due to any environmental abnormalities. The proteomic study revealed this kind of dehydration tolerance capability with the due course of their development [23][24]. In a nutshell, it can be suggested that the sulfate transporters SULTR3 and SULTR4 play a vital role in sulfate translocation, which is associated with seed development, by suppling sulfate and different S metabolites. In this context, it can also be noted that SULTR3 and SULTR4 homologs control the allocation of sulfate in between the seed compartments and help to modulate S metabolites and seed protein composition in Arabidopsis [23][24].
According to Zuber et al. [25], it was noted that different members of the SULTR3 family were more highly expressed at different stages of seed development of Arabidopsis than the other organs, which have the ability to control the translocation of sulfate within developing seeds. For instance, in developing embryos of chickpea (C. arietinum L.), various homologous members of the SULTR3 sub-type transporters were involved in sulfate transport and delivery as reported by Tabe et al. [26].
Another experiment depicted that, at bolting stage, previously stored sulfate in vacuole of the source leaves is remobilized into the developing seeds of oilseed rape (B. napus L.) and an up-regulated gene expression is noted of SULTR4;1 and SULTR4;2 transporters. These kinds of upregulated gene expression of two SULTR4-type transporters were also observed during the vegetative growth phase in old and mature leaves [27] and in roots [28] of oil seed rape (Brassica napus L.). In addition, Gironde et al. [29] revealed that at the reproductive stage, sulfate is the main source of S remobilized from the stored vacuolar sulfate of leaves via tonoplastic SULTR4-type transporters in S deficient condition. Moreover, Awazuhara et al. [20] revealed that sulfate transporter SULTR2.1 is also involved in the transfer of S into developing seeds of Arabidopsis. In addition, when the amount of abscisic acid (ABA) in freshly harvested seeds of sultr3;1 mutants was tested, 25–50% more ABA was found in comparison to wild-type Arabidopsis plants; this finding established the fact that SULTR3;1 affects ABA biosynthesis not only during the time of early vegetative growth but also in the seed filling stage [30].
4. Sulfur Regulated Seed Germination
Germination is the most important and the beginning phase of any plant’s life cycle. Seed germination is divided into three phases, during this course of time various metabolic activities take place including sulfate metabolism. Very little information is available regarding seed germination influenced by S-containing SSP and various S-regulated functional proteins and stored phytohormones present in seeds. Based on sulfate metabolism, seed germination was assessed by creating multigene defective mutants under abscisic acid (ABA) and salt conditions. In the same situation, a single mutation in SULTR3 transporters gene construct can alter seed germination responses. A progressive reduction in germination percentage was also observed from single to SULTR3 quintuple mutants, the latter showing the lowest rate of germination under both ABA and NaCl treatment [31]. After 5 days of sowing, the SULTR3 quintuple mutant showed only a 10% seed germination rate, whereas in the wild type, more than 60% germination was recorded under 0.3 mM ABA. Furthermore, at 8 days after sowing (DAS), only 20% of the SULTR3 quintuple mutant seeds were germinated, whereas in the wild type, more than 90% of the seeds were germinated. A similar pattern was observed under salt stress [31]. Consistent with alterations in Cys and ABA content, seed germination inhibition upon treatment with exogenous ABA and salt was directly proportional to the number of mutated SULTR3 genes, demonstrating the correlation of Cys and ABA content with the plant abiotic stress response during germination stages (Figure 1). In SULTR3 quadruple and quintuple mutants, delayed germination of seeds was noted under normal condition, in comparison to the wild type. In the case of triple mutants, sultr3;1 sultr3;2 sultr3;3, but not sultr3;2 sultr3;3 sultr3;4 or sultr3;3 sultr3;4 sultr3;5, showed a delayed germination at 3 DAS, which indicates that SULTR3;1 may play a more crucial role than other SULTR3 members during the early stage of seed germination of Arabidopsis [31].
Studies depicted that H2S is an essential component of sulfur and cysteine metabolism [32], is generated via different enzymatic and nonenzymatic mechanisms and acts as a signaling molecule in plant cells [33][34]. The metabolic process of H2S is dependent on the type of subcellular compartment and plant organ under optimal environmental conditions, as well as in stressful situation [33]. Seed physiological quality represented a higher rate of seed germination and longevity, lower reserves deterioration and higher seedling vigor. A reduced physiological quality of seeds decreases the germination percentage and vigor index of the seed [35], whereas the exposure of seeds in H2S under stress as well as normal conditions has generated positive effects on germination percentage and time, seedling growth, fresh weight, root length and other metabolic changes [36]. Moreover, Caverzan et al. [37] narrate that future studies are essential to understand the interaction between metabolic pathways related to H2S and how this signaling molecule is involved in improvement of seed germination process.
Germination is a very crucial process; here, a seed starts its second generation by coping with its new ambient conditions, which sometimes may create constrains for it. However, seed germination can be sped up via utilizing the knowledge of a new exciting technology, named seed priming, which is easy to handle, farmer friendly and helpful in sustaining nature, which led plant scientists to unveil a number of facts about the seeds via various omics-related studies and finally a new term also coined as “primeomics” [38]. However, Bose and Mishra [39] observed that only 24 h of pre-sowing soaking (one kind of seed priming technique) of mustard (B. rapa; a sulfur enriched crop) seeds with Mg(NO3)2 and MgSO4 salts improved the percent germination; furthermore, their finding validated that the affectivity can be carried over from vegetative phase to yield; this experiment depicted that NO3- and/or SO42- might be a responsible factor for the improvement of yield. This may give support to the presence of low affinity sulfate transporters in seeds which may have worked effectively in the presence of MgSO4 solution, during the process of priming of mustard seeds [40][41].
This entry is adapted from the peer-reviewed paper 10.3390/plants11030450
References
- Hawkesford, M.J. Sulfur. In Nutritional Genomics; Broadley, M.R., White, P., Eds.; Blackwell Publishers: Oxford, UK, 2005; pp. 87–111.
- Abou-Seeda, M.A.; Abou Ei-Nour, E.A.A.; Yassen, A.A.; Gad Mervat, M.; Sahar, M.Z. Importance of sulfur and its roles in plants physiology—A review. Current Sci. Int. 2020, 9, 198–231.
- Li, Q.; Gao, Y.; Yang, A. Sulfur homeostasis in plants. Int. J. Mol. Sci. 2020, 21, 8926.
- Smith, F.W.; Ealing, P.M.; Hawkesford, M.J.; Clarkson, D.T. Plant members of a family of sulfate transporters reveal functional subtypes. Proc. Natl. Acad. Sci. USA 1995, 92, 9373–9377.
- Noctor, G.L.; Gomez, H.; Vanacker; Foyer, C.H. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signaling. J. Exp. Bot. 2002, 53, 1283–1304.
- Lunde, C.; Zygaldo, A.; Simonsen, H.T.; Nielsen, P.L.; Blennow, A.; Haldrup, A. Sulfur starvation in rice: The effect on photosynthesis, carbohydrate metabolism, and oxidative stress protective pathways. Physiol. Plant. 2008, 134, 508–521.
- Tabe, L.M.; Droux, M. Sulfur assimilation in developing lupin cotyledons could contribute significantly to the accumualtion of organic sulfur reserves in seed. Plant Physiol. 2001, 126, 176–187.
- Martinoia, E.; Maeshima, M.; Neuhaus, H.E. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007, 58, 83–102.
- Martinoia, E.; Massonneau, A.; Frangne, N. Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol. 2000, 41, 1175–1186.
- Gallardo, K.; Firnhaber, C.; Zuber, H.; He´richer, D.; Belghazi, M.; Henry, C.; Küster, H.; Thompson, R. A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: Evidence for metabolic specialization of maternal and filial tissues. Mol. Cell Proteom. 2007, 6, 2165–2179.
- Takahashi, H. Sulphate transport system in plants: Functional diversity and molecular mechanisms underlying regulatory coordination. J. Exp. Bot. 2019, 70, 475–4087.
- Rouached, H.; Berthomieu, P.; El Kassis, E.; Cathala, N.; Catherinot, V.; Labesse, G.; Davidian, J.C.; Fourcroy, P. Structural and Functional Analysis of the Cterminal STAS (Sulfate Transporter and Anti-sigma Antagonist) Domain of the Arabidopsis thaliana Sulfate Transporter SULTR1. 2. J. Biol. Chem. 2005, 280, 15976–15983.
- Buchner, P.; Stuiver, E.; Westerman, S.; Wirtz, M.; Hell, R.; Hawkesford, M.; De Kok, L. Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol. 2004, 136, 3396–3408.
- Tomatsu, H.; Takano, J.; Takahashi, H.; Watanabe-Takahashi, A.; Shibagaki, N.; Fujiwara, T. An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. USA 2007, 104, 18807–18812.
- El Kassis, E.; Cathala, N.; Rouached, H.; Fourcroy, P.; Berthomieu, P.; Terry, N.; Davidian, J.C. Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiol. 2007, 143, 1231–1241.
- Shibagaki, N.; Rose, A.; McDermott, J.P.; Fujiwara, T.; Hayashi, H.; Yoneyama, T.; Davies, J.P. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002, 29, 475–486.
- Smith, F.W.; Hawkesford, M.J.; Ealing, P.M.; Clarkson, D.T.; Vanden Berg, P.J.; Belcher, A.R.; Warrilow, A.G. Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J. 1997, 12, 875–884.
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The roles of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182.
- Yoshimoto, N.; Takahashi, H.; Smith, F.W.; Yamaya, T.; Saito, K. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002, 29, 465–473.
- Awazuhara, M.; Fujiwara, T.; Hayashi, H.; Watanabe-Takahashi, A.; Takahashi, H.; Saito, K. The function of SULTR2;1 sulfate transporter during seed development in Arabidopsis thaliana. Physiol. Plant. 2005, 125, 95–105.
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot transport of sulfate in Arabidopsis: Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004, 136, 4198–4204.
- Kataoka, T.; Watanabe-Takahashi, A.; Hayashi, N.; Ohnishi, M.; Mimura, T.; Buchner, P.; Hawkesford, M.J.; Yamaya, T.; Takahashi, H. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 2004, 16, 2693–2704.
- Zuber, H.; Davidian, J.C.; Aubert, G.; Aimé, D.; Belghazi, M.; Lugan, R.; Heintz, D.; Wirtz, M.; Hell, R.; Thompson, R.; et al. The seed composition of Arabidopsis mutants for the group3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol. 2010, 154, 913–926.
- Zuber, H.; Davidian, J.-C.; Wirtz, M.; Hell, R.; Belghazi, M.; Thompson, R.; Gallardo, K. Sultr4;1 mutant seeds of Arabidopsis have an enhanced sulfate content and modified proteome suggesting metabolic adaptations to altered sulfate compartmentalization. BMC Plant Biol. 2010, 10, 78.
- Zuber, H.; Aubert, G.; Davidian, J.-C.; Thompson, R.; Gallardo, K. Sulfur metabolism and transport in developing seeds. In Sulfur metabolism in Plants; Sirko, A., De Kok, L.J., Haneklaus, S., Hawkesford, M.J., Rennenberg, H., Saito, K., Schnug, E., Stulen, I., Eds.; Backhuys Publishers: Leiden, The Netherlands, 2009; pp. 113–118.
- Tabe, L.M.; Venables, I.; Grootemaat, A.; Lewis, D. Sulfur transport and assimilation in developing embryos of chickpea (Cicer arietinum). In Sulfur Transport and Assimilation in Plants; Davidian, J.-C., Grill, D., De Kok, L.J., Stulen, I., Hawkesford, M.J., Schnug, E., Rennenberg, H., Eds.; Backhuys Publishers: Leiden, The Netherlands, 2003; pp. 335–337.
- Dubousset, L.; Abdallah, M.; Desfeux, A.S.; Etienne, P.; Meuriot, F.; Hawkesford, M.J.; Gombert, J.; Bonnefoy, J.; Ameline, A.F.; Ourry, A.; et al. Remobilization of leaf S compounds and senescence in response to restricted sulfate supply during the vegetative stage of oilseed rape are affected by mineral N availability. J. Exp. Bot. 2009, 60, 3239–3253.
- Parmar, S.; Buchner, P.; Hawkesford, M.J. Leaf developmental stage affects sulfate depletion and specific sulfate transporter expression during sulfur deprivation in Brassica napus L. Plant Biol. 2007, 9, 647–653.
- Gironde, A.; Dubousset, L.; Trouverie, J.; Etienne, P.; Avice, J.-C. The impact of sulfate restriction on seed yield and quality of winter oil seed rape depends on the ability to remobilize sulfate from vegetative tissues to reproductive organs. Front. Plant Sci. 2014, 5, 695.
- Cao, M.-J.; Wang, Z.; Zhao, Q.; Mao, J.-L.; Speiser, A.; Wirtz, M.; Hell, R.; Zhu, J.-K.; Xiang, C.-B. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. The Plant J. 2014, 77, 604–615.
- Chen; et al SULTR3s function in chloroplast sulphate uptake and affect ABA biosynthesis & the stress response. Plant Physiol. 2019, 180, 593–604.
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of storage proteins in plant seeds is mediated by amyloid formation. PLoS Biol. 2020, 18, e3000564.
- Chen, Z.; Zhao, P.; Miao, Z.; Qi, G.; Wang, Z.; Yuan, Y.; Ahmad, N.; Cao, M.; Hell, R.; Wirtz, M.; et al. Hydrogen Sulfide: A New Warrior against Abiotic Stress. Trends. Plant Sci. 2019, 24, 983–988.
- Huo, J.; Huang, D.; Zhang, J.; Fang, H.; Wang, B.; Wang, C.; Liao, W. Hydrogen sulfide: A gaseous molecule in postharvest freshness. Front. Plant Sci. 2018, 9, 1172.
- Ebone, L.A.; Caverzan, A.; Chavarria, G. Physiologic alterations in orthodox seeds due to deterioration processes. Plant Physiol. Biochem. 2019, 145, 34–42.
- Corpas, F.J.; Palma, J.M. H2S signaling in plants and applications in agriculture. J. Adv. Res. 2020, 24, 131–137.
- Caverzan, A.; Ebone, L.A.; Chiomento, J.L.T.; Chavarria, G. Physiological role of hydrogen sulfide in seed germination and seedling development under stress conditions. Arch. Agron. Soil Sci. 2021.
- Srivastava, A.K.; Kumar, J.S.; Suprasanna, P. Seed ‘primeomics’: Plants memorize their germination under stress. Biol. Rev. Camb. Philos. Soc. 2021, 96, 1723–1743.
- Bose, B.; Mishra, T. Effect of Mg-salts on seed germination of Brassica sp. Cruciferae New Lett. 1997, 19, 63–64.
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2011, 2, 119.
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184.
This entry is offline, you can click here to edit this entry!
