Secreted non-coding RNAs play crucial roles during cancer progression and strongly contribute to remodel the tumor microenvironment and the metastatic niche, to enable the formation of a supporting vasculature, the inhibition of tumor recognition by the immune system and, finally, the spreading of tumor cells and metastatization. The full comprehension of the ncRNA-guided networks at the basis of these events is central for the development of novel effective therapies aimed at disrupting the cross-talk between tumor cells and other cell types in the tumor microenvironment; such therapeutic approaches would strongly prompt the immune system to recognize and eliminate tumor cells. At the same time secreted non-coding RNAs also represent powerful biomarkers to be exploited for diagnostic in liquid biopsy and for therapeutic purposes. A comprehensive understanding of the mechanisms of action of secreted ncRNAs in TNBC represents the future challenge, which will allow the widest use of these molecules both as diagnostic tools and as therapeutic targets.
- breast cancer
- TNBC
- non-coding RNA
- ncRNA
- exosomes
- tumor microenvironment
- liquid biopsy
1. Introduction
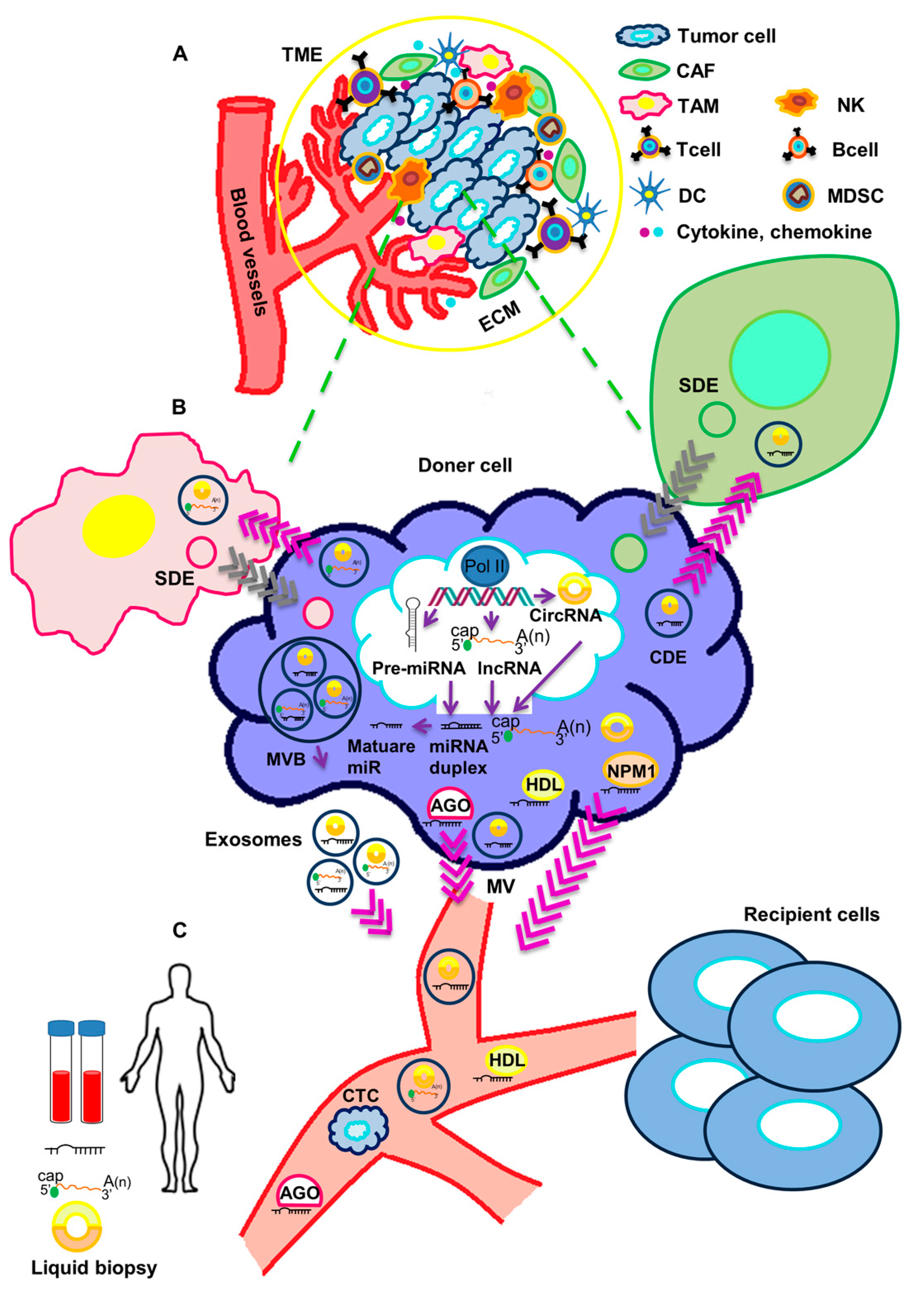 Figure 1. Crosstalk of circulating non-coding RNAs and tumor microenvironment. (A) The tumor microenvironment (TME) includes a cellular and an extracellular component. The cellular component consists of tumor cells, immune cells (TAMs, T cells, B cells, NK cells, DCs, and MDSCs) and cancer-associated fibroblasts (CAFs). The extracellular component of TME is composed by ECM proteins and signaling molecules (like cytokines, chemokines, growth factors, hormones, etc.), which are secreted by the cellular component. (B) ncRNAs are processed and released in body fluids. Circulating ncRNAs can be released in two ways: (1) as cell-free RNAs, complexed with protein such as AGO, NPM1 or HDL; or (2) in extracellular vesicles (exosomes and microvesicles). Both tumor cells and stromal cells may release exosomes in the TME. Through body fluids, ncRNAs can reach distant sites in the body and act as molecular mediators. (C) Circulating ncRNAs are stable and easily detectable and can be used as non-invasive biomarkers in liquid biopsy. TME: tumor microenvironment, TAM: Tumor-associated macrophage, NK: Natural killer cell, DC: Dendritic cell, MDSC: Myeloid-derived suppressor cell, CAF: cancer-associated fibroblast, ECM: extracellular matrix, SDE: Stromal-derived exosome, CDE: Cancer-derived exosome, AGO: Argonaute2, NPM1: Nucleophosmin 1, HDL: High-density lipoprotein, MVB: Multivesicular body, MV: Microvesicle, CTC: Circulating tumor cell.
Figure 1. Crosstalk of circulating non-coding RNAs and tumor microenvironment. (A) The tumor microenvironment (TME) includes a cellular and an extracellular component. The cellular component consists of tumor cells, immune cells (TAMs, T cells, B cells, NK cells, DCs, and MDSCs) and cancer-associated fibroblasts (CAFs). The extracellular component of TME is composed by ECM proteins and signaling molecules (like cytokines, chemokines, growth factors, hormones, etc.), which are secreted by the cellular component. (B) ncRNAs are processed and released in body fluids. Circulating ncRNAs can be released in two ways: (1) as cell-free RNAs, complexed with protein such as AGO, NPM1 or HDL; or (2) in extracellular vesicles (exosomes and microvesicles). Both tumor cells and stromal cells may release exosomes in the TME. Through body fluids, ncRNAs can reach distant sites in the body and act as molecular mediators. (C) Circulating ncRNAs are stable and easily detectable and can be used as non-invasive biomarkers in liquid biopsy. TME: tumor microenvironment, TAM: Tumor-associated macrophage, NK: Natural killer cell, DC: Dendritic cell, MDSC: Myeloid-derived suppressor cell, CAF: cancer-associated fibroblast, ECM: extracellular matrix, SDE: Stromal-derived exosome, CDE: Cancer-derived exosome, AGO: Argonaute2, NPM1: Nucleophosmin 1, HDL: High-density lipoprotein, MVB: Multivesicular body, MV: Microvesicle, CTC: Circulating tumor cell.2. Functional Impact of Secreted ncRNAs on Surrounding Stromal Cells and at Metastatic Sites in TNBC
This entry is adapted from the peer-reviewed paper 10.3390/ncrna8010005
References
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690.
- Lee, A.; Djamgoz, M.B.A. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat. Rev. 2018, 62, 110–122.
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61.
- Nedeljkovic, M.; Damjanovic, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019, 8, 957.
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310.
- Zhang, Z.; Zhang, J.; Diao, L.X.; Han, L. Small non-coding RNAs in human cancer: Function, clinical utility, and characterization. Oncogene 2021, 40, 1570–1577.
- Ono, S.; Lam, S.; Nagahara, M.; Hoon, D.S.B. Circulating microRNA Biomarkers as Liquid Biopsy for Cancer Patients: Pros and Cons of Current Assays. J. Clin. Med. 2015, 4, 1890–1907.
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563.
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170.
- Tang, G.L.; Tang, X.Q.; Mendu, V.; Tang, X.H.; Jia, X.Y.; Chen, Q.J.; He, L.H. The art of microRNA: Various strategies leading to gene silencing via an ancient pathway. Bba-Gene Regul. Mech. 2008, 1779, 655–662.
- Takahashi, R.; Prieto-Vila, M.; Kohama, I.; Ochiya, T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019, 110, 1140–1147.
- Tessitore, A.; Cicciarelli, G.; Mastroiaco, V.; Del Vecchio, F.; Capece, D.; Verzella, D.; Fischietti, M.; Vecchiotti, D.; Zazzeroni, F.; Alesse, E. Therapeutic Use of MicroRNAs in Cancer. Anti-Cancer Agent Me 2016, 16, 7–19.
- Blandino, G.; Fazi, F.; Donzelli, S.; Kedmi, M.; Sas-Chen, A.; Muti, P.; Strano, S.; Yarden, Y. Tumor suppressor microRNAs: A novel non-coding alliance against cancer. Febs. Lett. 2014, 588, 2639–2652.
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651.
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Geng, X.C.; Jia, Y.C.; Zhang, Y.H.; Shi, L.; Li, Q.; Zang, A.M.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinsuogens. Epigenomics 2020, 12, 267–283.
- Zhao, W.S.; Dong, M.; Pan, J.R.; Wang, Y.J.; Zhou, J.Y.; Ma, J.J.; Liu, S.Y. Circular RNAs: A novel target among non-coding RNAs with potential roles in malignant tumors. Mol. Med. Rep. 2019, 20, 3463–3474.
- Fontemaggi, G.; Turco, C.; Esposito, G.; Di Agostino, S. New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers 2021, 13, 3154.
- Bullock, M.D.; Silva, A.M.; Kanlikilicer-Unaldi, P.; Filant, J.; Rashed, M.H.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. Exosomal Non-Coding RNAs: Diagnostic, Prognostic and Therapeutic Applications in Cancer. Noncoding RNA 2015, 1, 53–68.
- Cui, M.Y.; Wang, H.D.; Yao, X.X.; Zhang, D.; Xie, Y.J.; Cui, R.J.; Zhang, X.W. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626.
- Yan, X.Q.; Xie, Y.H.; Yang, F.; Hua, Y.J.; Zeng, T.Y.; Sun, C.X.; Yang, M.Z.; Huang, X.; Wu, H.; Fu, Z.Y.; et al. Comprehensive description of the current breast cancer microenvironment advancements via single-cell analysis. J. Exp. Clin. Canc. Res. 2021, 40, 142.
- Chen, Q.; Li, Y.F.; Liu, Y.Q.; Xu, W.L.; Zhu, X.L. Exosomal Non-coding RNAs-Mediated Crosstalk in the Tumor Microenvironment. Front. Cell Dev. Biol 2021, 9, 646864.
- Baghba, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59.
- Wang, S.E. Extracellular Vesicles and Metastasis. Csh. Perspect. Med. 2020, 10, a037275.
- Pigati, L.; Yaddanapudi, S.C.S.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective Release of MicroRNA Species from Normal and Malignant Mammary Epithelial Cells. PLoS ONE 2010, 5, e13515.
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721.
- Madhavan, D.; Zucknick, M.; Wallwiener, M.; Cuk, K.; Modugno, C.; Scharpff, M.; Schott, S.; Heil, J.; Turchinovich, A.; Yang, R.X.; et al. Circulating miRNAs as Surrogate Markers for Circulating Tumor Cells and Prognostic Markers in Metastatic Breast Cancer. Clin. Cancer Res. 2012, 18, 5972–5982.
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncology 2012, 14, 689–700.
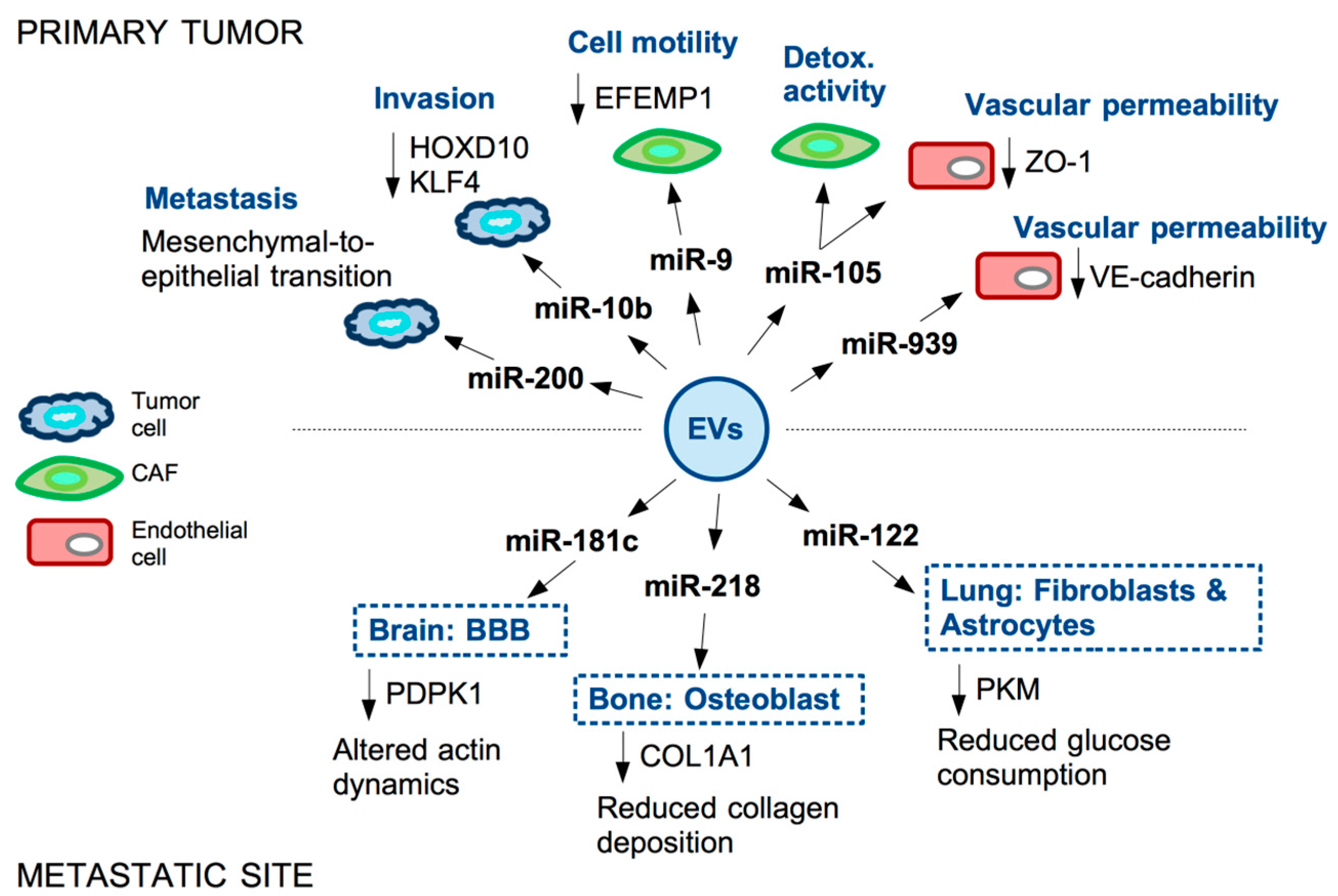
- Le, M.T.N.; Hamar, P.; Guo, C.Y.; Basar, E.; Perdigao-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128.
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.H.; Mo, Y.Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256.
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186.
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–252.
- Baroni, S.; Romero-Cordoba, S.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016, 7, e2312.
- Cosentino, G.; Romero-Cordoba, S.; Plantamura, I.; Cataldo, A.; Iorio, M.V. miR-9-Mediated Inhibition ofEFEMP1Contributes to the Acquisition of Pro-Tumoral Properties in Normal Fibroblasts. Cells 2020, 9, 2143.
- Yan, W.; Wu, X.W.; Zhou, W.Y.; Fong, M.Y.; Cao, M.H.; Liu, J.; Liu, X.J.; Chen, C.H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 2018, 20, 597–609.
- Zhou, W.Y.; Fong, M.Y.; Min, Y.F.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515.
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100.
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lotvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716.
- Liu, X.X.; Cao, M.H.; Palomares, M.; Wu, X.W.; Li, A.; Yan, W.; Fong, M.Y.; Chan, W.C.; Wang, S.E. Metastatic breast cancer cells overexpress and secrete miR-218 to regulate type I collagen deposition by osteoblasts. Breast Cancer Res. 2018, 20, 127.
- Fong, M.Y.; Zhou, W.Y.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194.
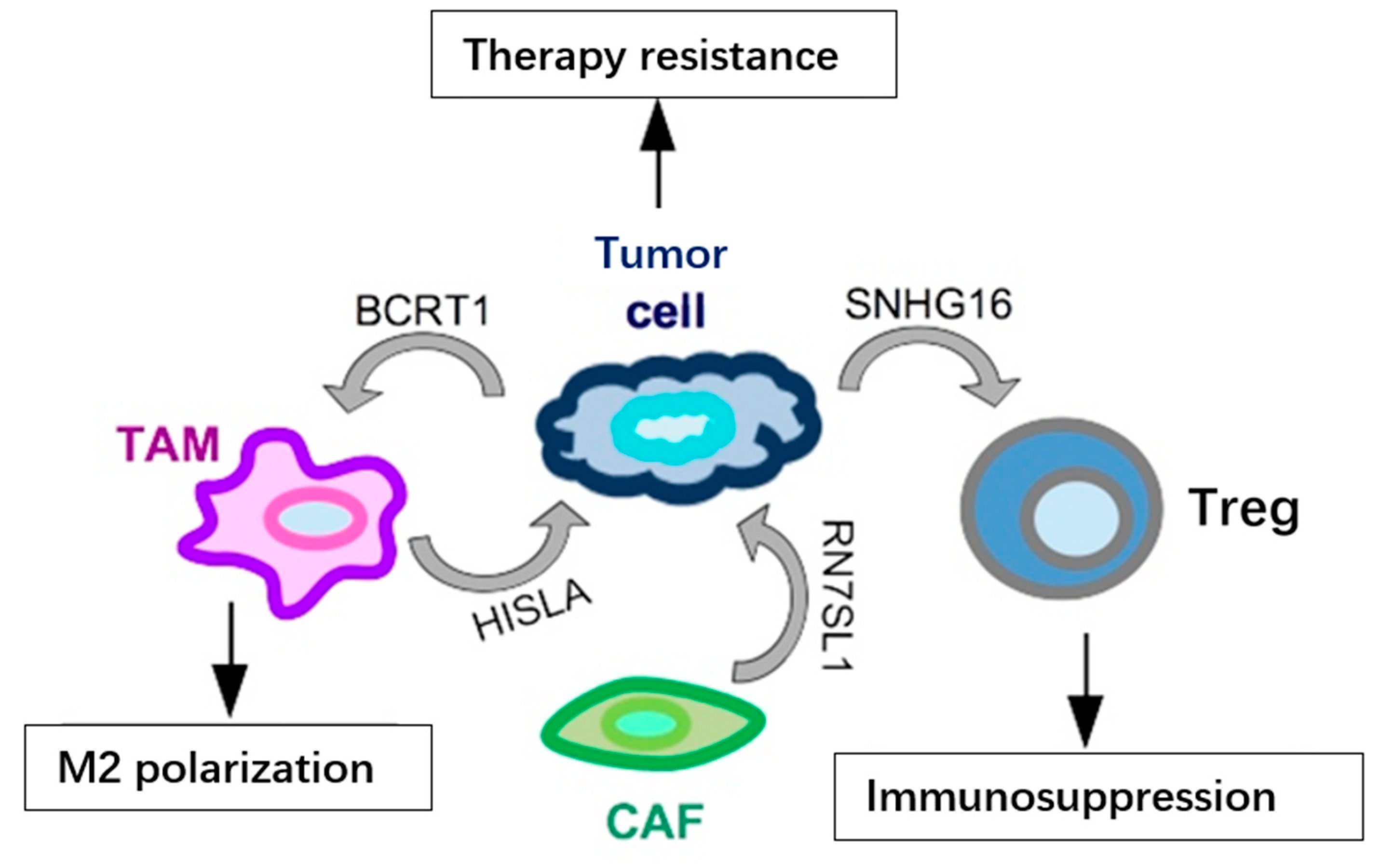
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+gamma delta 1 Treg cells. Signal. Transduct Tar. 2020, 5, 41.
- Liang, Y.R.; Song, X.J.; Li, Y.M.; Chen, B.; Zhao, W.J.; Wang, L.J.; Zhang, H.W.; Liu, Y.; Han, D.W.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 127.
- Nabet, B.Y.; Qiu, Y.; Shabason, J.E.; Wu, T.J.; Yoon, T.; Kim, B.C.; Benci, J.L.; DeMichele, A.M.; Tchou, J.; Marcotrigiano, J.; et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell 2017, 170, 352–366.
- Chen, F.; Chen, J.N.; Yang, L.B.; Liu, J.; Zhang, X.Q.; Zhang, Y.; Tu, Q.Q.; Yin, D.; Lin, D.C.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1 alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510.
- Zhang, P.; Zhou, H.X.; Lu, K.F.; Lu, Y.N.; Wang, Y.; Feng, T.B. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Oncotargets Ther. 2018, 11, 291–299.
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63.
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Khattak, S.; Alajez, N.M. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Discov. 2021, 7, 23.
- Pruszko, M.; Milano, E.; Forcato, M.; Donzelli, S.; Ganci, F.; Di Agostino, S.; De Panfilis, S.; Fazi, F.; Bates, D.O.; Bicciato, S.; et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. Embo. Rep. 2017, 18, 1331–1351.
