Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Circular RNAs (circRNAs) are an emerging group of long non-coding RNAs (lncRNAs) and have attracted attention again according to the progress in high-throughput sequencing in recent years. circRNAs are genome transcripts produced from pre-messenger (m)RNA regions in a specific process called “back-splicing,” which forms covalently closed continuous loops.
- circRNA
- obesity
- diabetes
- NAFLD
- NASH
1. Introduction
Statistics from the World Health Organization from 2000 to 2019 reveal that “non-communicable diseases” accounted for seven of the top 10 causes of death worldwide. Heart disease still ranks first among all diseases [1]. Diabetes is also among the top 10, and the related death toll increased during that period [1]. This phenomenon reflects changes in human lives in modern times. By analyzing risk factors of cardiovascular disease and diabetes, one can find many similarities, with the most notorious one being metabolic syndrome (MetS). MetS describes a clustering of the dysregulation of several metabolic processes, including hyperglycemia, hyperlipidemia, and abnormal adipose deposition. Per the concept of syndrome X, as proposed by Dr. Reaven, a pioneer in metabolic syndrome research, this clustering phenomenon is closely related to insulin resistance (IR) [2].
IR refers to the phenomenon of an insufficient response by fat cells, muscle cells, and liver cells to normal circulatory levels of insulin [3]. Several major mechanisms were suggested, including oxidative stress, inflammation, insulin receptor mutations, endoplasmic reticular (ER) stress, and mitochondrial dysfunction.
2. The Biogenesis, Biology, and Characterization of circRNAs
Viroids, which are small, circular, single-stranded RNA molecules (of 246–401 nucleotides) that are uncoated and pathogenic to higher plants, were the first circRNA molecules to be discovered in 1976 [4]. A few years later, circRNAs were observed in the cytoplasmic fractions of eukaryotic cell lines by electron microscopy. However, due to a lack of suitable research tools, circRNAs were once considered to be “junk” products produced by aberrant RNA splicing. They have low abundances in cells and low sequence conservation, and received little attention until 1993, when it was discovered that Sry, the sex-determining gene, undergoes circular transcription in adult mouse testicles [5].
Recently, with advances in high-throughput sequencing (Seq) technologies, Salzman reported ~80 circular RNAs for the first time through the RNA-Seq method [6]. Since then, large numbers of circRNA molecules have been discovered. Jeck et al. detected more than 25,000 circRNAs in human fibroblasts; Memczak et al. identified 1950 human circRNAs and 1903 mouse species through RNA-Seq data combined with a human leukocyte database (81 of which were the same as human circRNAs) and 724 nematode circRNAs [7][8]. Of note, with these new discoveries, people began to recognize that circRNAs are abundant, diverse, and conserved molecules in tissue- and developmental stage-specific manners and might play critical roles in human biological processes [6][7][8].
2.1. The Biogenesis of circRNAs
Although it was proposed that circRNAs are derived from precursor messenger (m)RNAs (pre-mRNAs) by back-splicing, the mechanism of circRNA biogenesis still remains elusive. circRNAs are generally generated by joining an upstream 3′ splice site to a downstream 5′ splice site to form a covalently closed loop [9]. Based on the original sequences, circRNAs can mainly be classified in three categories: exonic circular (ecirc)RNAs [7][8][10], circular intronic (ci)RNAs [11], and exon–intron circular (EIci)RNAs [12]. The majority (over 80%) of circRNAs mainly reside in the cytoplasm, while ciRNAs and EIciRNAs are mainly found in nuclei [7][8][10]. Briefly, the formation of circRNAs can be classified into two popular models (Figure 1), the direct back-splicing model and the lariat precursor model.
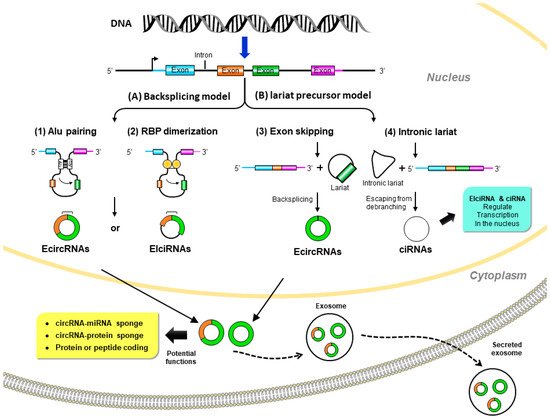
Figure 1. Biogenesis and potential biological functions of circular (circ)RNAs. (A) Back-splicing model: pre-mRNA is spliced in a non-canonical manner of “back-splicing” by (1) inverted repeat elements in long flanking intron pairs, such as Alu elements, or (2) dimerization of RNA-binding proteins (RBPs). During back-splicing, an upstream branch point attacks a downstream splice donor site to form exonic circRNAs or exon–intron circRNAs. (B) Lariat precursor model: pre-mRNA undergoes canonical splicing to generate a linear mRNA and a lariat precursor. (3) The lariat precursors with exon components might be generated from exon-skipping events and then further back-spliced to exonic circRNAs. Alternatively, (4) the intronic lariat precursors escape from the debranching step of canonical linear splicing to form intronic circRNAs. Exonic circRNAs are transported from the nucleus to the cytoplasm to function as miRNA sponges to inhibit miRNA activity; protein sponges (such as RBPs) affect protein functions and translocation or protein-coding to further translation. Exon–intron circRNAs and intronic circRNAs can interact with transcription complexes to regulate transcription in the nucleus.
2.1.1. Direct Back-Splicing Model
In this model, paired elements flanking the downstream and upstream splices bring these sites into proximity to form a loop structure that favors back-splicing. Intron pairing, such as Alu elements [13], or the dimerization of RNA-binding proteins (RBPs), such as Muscleblind (MBL) [14] and Quaking (QKI) [15], initiate circularization in circRNA formation.
2.1.2. Lariat Precursor Model
In addition to the back-splicing mechanism, circRNAs can also be produced from the processing of lariat sequences. Standard linear splicing converts an intron into a looped structure called a ‘lariat’ (like a lasso) [16]. The splicing machinery then generally eliminates the lariat. This mode includes the following two ways: (a) exon-skipping: during alternative linear splicing, a giant lariat containing the skipped exon can originate. These exon-containing lariats may further form mature circRNAs by back-splicing [17][18]. (b) Intron lariat: pre-mRNAs occur due to canonical linear splicing to from linear mRNA and a long intron lariat. Then the intron lariat that escaped from debranching is further back-spliced to produce mature circRNA [11].
Although the production of circRNA is still a mystery, the production efficiency is very low due to the sterically unfavorable connections between the downstream 5’ splice site and the upstream 3‘ splice site [19]. Nevertheless, because they are resistant to RNA exonucleases or RNase R, circRNAs can accumulate at higher concentration than linear RNAs in quiescent and post-mitotic cells, such as neurons [20]. At the same time, circRNAs are also rich in exosomes and could be found in extracellular fluid (like blood) at levels higher than linear RNA [21]. Therefore, circRNAs are considered good biomarkers for disease diagnosis.
2.2. Functions of circRNAs
The main functions of circRNAs are still not well understood, but growing data suggest that they play lots of essential roles in many biological processes. People proposed that circRNAs in different locations have diverse functions. While cytoplasmic circRNAs sponge miRNAs or proteins, nuclear circRNAs regulate transcription and alternative splicing events.
2.2.1. circRNAs Act as miRNA Sponges
In 2013, two research teams published the first observations that circRNAs can indirectly modulate gene expressions at the posttranscriptional level by sequestering miRNA and mRNA interactions [6][7]. Those studies also noted that ciRS-7 circRNA contained more than 70 conserved binding sites for the miR-7 sponge, thereby regulating expression of miR-7 target mRNAs. Interestingly, ciRS-7-knockout mice had lower miR-7 levels, impaired sensorimotor gating, and enhanced miR-7 target genes, such as Fos, an immediate early gene [22]. That in vivo model suggested that interactions between ciRS-7 and miRNAs are important for essential brain functions.
Of note, ciRS-7’s inhibiting or protecting miR-7 from degradation may vary with the cellular context [22][23][24][25]. Finally, many other circRNAs, including circHIPK3 [26] and circBIRC6 [27], were shown to have miRNA sponging abilities. However, unlike ciRS-7, these circRNAs contain fewer miRNA binding sites than expected by chance [28].
2.2.2. circRNAs Interact with Proteins
circRNAs can serve as protein decoys to regulate the function of RBPs, for example, circRNA poly(A)-binding protein nuclear 1 (circPABPN1) and circular antisense noncoding RNA in the INK4 locus (circANRIL). CircPABPN1 suppresses the translation of nuclear poly(A)-binding protein 1 (PABPN1) mRNA by sequestering the protein RBP Hu-antigen R (HUR) [29]. In vascular smooth muscle cells and macrophages, circANRIL sequesters pescadillo homologue 1 (PES1), an essential 60S preribosomal assembly factor, to impair rRNA maturation, resulting in atherosclerosis-related nucleolar stress, p53 activation, and cell apoptosis [30]. circRNAs can also function as protein scaffolds. circFoxo3 combined with the cell cycle proteins, cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21), form the circFoxo3–p21–CDK2 ternary complex and inhibit CDK2 function to repress cell cycle progression [31]. Recently, extensive screening of circRNA-RBP interactions was performed, and the concept of a circRNA-RBP interactome raises the emergent crucial regulatory roles in tumorigenesis and other vital cellular functions [32].
2.2.3. circRNAs as Regulators of Transcription and Splicing
Recently, it was revealed that in the nucleus, both ciRNAs and EIciRNAs can modulate their parental gene transcription by interacting with upstream promoters, RNA polymerase II (Pol II), and other transcription machinery proteins [11][12][33]. Two ciRNAs, ci-ANKRD52 and ci-SIRT-7, enhance their parental gene expressions of ankyrin repeat domain 52 (ANKRD52) and sirtuin 7 (RIRT7), respectively [11][33]. By contrast, EIciRNAs, such as circEIF3J and circPAIP2, interact with the U1 small nuclear ribonucleoprotein (U1 snRNP) and RNA polymerase II (Pol II) via U1 snRNA-binding sites, thereby enhancing expressions of their parent genes [12].
Certain circRNAs also act on gene expressions trans-functionally by competing with linear splicing. For example, circMbl was proposed to function as a protein sponge, and it originates from the gene that encodes the splicing factor, muscleblind (mbl), in Drosophila melanogaster and the homologous gene, muscleblind-like protein 1 (MBNL1), in humans [12]. Interestingly, mbl itself directly regulates the biogenesis of circMbl by the presence of specific mbl-binding sites in the introns flanking the circularizable exon (also shown in MBNL) [12]. Therefore, this autoregulatory circuit may result in excess mbl or MBNL1 decreasing the production of its mRNA by promoting circMbl biogenesis, and the circMbl helps the linear splicing of the gene by tethering mbl or MBNL1 [12].
2.2.4. circRNAs Translate to Proteins
Although circRNAs do not contain 5‘ caps or poly(A) tails, it was shown that circRNAs are translated in a cap-independent manner by the internal ribosome entry site (IRES) [34] and N6-methyl-adenosines (m6A) [35][36]. circ-ZNF609 was the first to be identified as a circRNA that makes a protein function involved in muscle development [37]. Additionally, the circular form of the long intergenic non-protein-coding RNA p53-induced transcript (LINC-PINT) can be translated into a small peptide involved in glioblastoma tumorigenesis [38]. Although there are many examples in the literature of circRNAs being translated into proteins or peptides, the functions of those molecules remain unknown.
3. circRNAs Regulate Insulin Signaling and β-Cell Function
Pancreatic β-cells are the only source of insulin in the body and play a major role in glucose homeostasis. Type 1 diabetes, which mostly occurs in adolescents and children, is mainly caused by autoimmune problems, destroying pancreatic β-cells, leading to a lack of insulin and requiring insulin injections. In contrast, the main reason for type 2 diabetes (T2D) is IR due to dysfunction in insulin signaling and subsequent hyperglycemic status. The pancreas secretes more insulin to allow blood sugar to enter cells but the compensatory increase in insulin is limited. Over time, prolonged hyperglycemia leads to β-cell dysfunction, proliferation impairment, and apoptosis activation, forming a vicious circle of β-cell exhaustion. There is emerging evidence that circRNAs are involved in regulating β-cell functions and in diabetes development (Figure 2) [39][40][41][42].
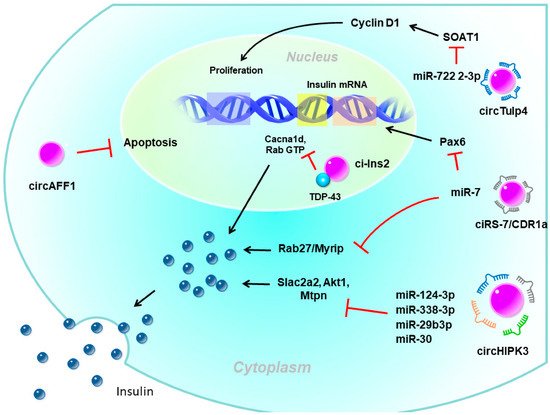
Figure 2. The potential role of circular (circ)RNAs in pancreatic β-cells. circHIPK3 and ciRS-7/CDR1a are exonic circRNAs mainly localized in the cytoplasm which act as miRNA sponges to enhance insulin secretion. ciRS-7/CDR1a also enhances insulin expression by the mi7 sponge. circTulp4 is also an exonic circRNA that sequesters miR-7222-3p to promote β-cell proliferation. circAFF1 is an exonic circRNA that inhibits β-cell apoptosis through an unknown mechanism. ci-Ins2 is an intronic circRNA mainly localized in the nucleus that interacts with the RNA-binding protein, TDP-43. The interaction between ci-Ins2 and TDP-43 promotes expression of the insulin secretory machinery. Pax6, paired box 6; SLc2a2, solute carrier family 2 member 2; Akt1, AKT serine/threonine kinase 1; Mtpn, myotrophin; Myrip, myosin VIIA and Rab interacting protein; Cacna1d, calcium voltage-gated channel subunit alpha1 D; TDP-43, TAR DNA-binding protein 43.
3.1. circRNAs Regulate Insulin Secretion and β-Cell Function
circRNAs have been proposed to be involved in the regulation of insulin secretion. The most well-known circRNAs with such functions were ciRS-7/CDR1a and circHIPK3. CDR1as, which originates from the antisense transcript of the cerebellar degeneration-related protein 1 gene, was the first circRNA studied in pancreatic β-cells [43][44]. Since it contains more than 70 binding sites for miR-7, it is also named ciRS-7 (circRNA sponge for miR-7) [43][44]. Studies showed that miR-7 is one of the most common miRNAs expressed by β-cells and is a negative regulator of glucose-stimulated insulin secretion (GSIS) in β-cells [45]. Interestingly, while the overexpression of miR-7 in transgenic mouse β-cells causes diabetes due to impaired insulin secretion [45], overexpression of ciRS-7 leads to increased insulin secretion by islet cells by upregulating miR-7 target gene expressions, such as Myrip (myosin VIIA and Rab-interacting protein) [44]. ciRS-7 also enhances insulin transcription by regulating Pax6 (paired box 6) expression after sponging for miR-7 [44].
Furthermore, circHIPK3 is another very abundant islet circRNA and was reported to be reduced in islets of db/db mice [40]. Silencing of circHIPK3 caused defective insulin secretion, increased apoptosis, and reduced proliferation in MIN6B1 cells [40]. Furthermore, expressions of critical insulin secretion genes, such as Slc2a2, Akt1, and Mtpn, were downregulated upon circHIPK3 silencing [40]. Mechanistic studies proposed that these effects of circHIPK3 may be mediated via miRNA sponging, such as miR-124-3p, miR-338-3p, miR-29b-3p, and miR-30, which have known functions in β-cells [40]. Nevertheless, circHIPK3 silencing resulted in a decrease in the activity of a luciferase construct containing the 3‘ untranslated region (UTR) of human MTPN, which is known to be controlled by miR-124-3p [40].
Recently, ci-Ins2, derived from intron 2 of preproinsulin 2 (Ins2), also was suggested to be involved in insulin secretion. Silencing rat ci-Ins2 or ci-INS (the human homolog of ci-Ins2) was associated with reduced mRNA expressions that encode vital components of the insulin secretory machinery, including the voltage-dependent Ca2+ channel subunit, Cacna1d, and different targets and regulators of Rab3 GTPases [46]. TAR DNA-binding protein 43 (TDP-43)-knockout and ci-Ins2 silencing shared a common set of downregulated genes, especially genes involved in insulin exocytosis [47]. Interestingly, the level of ci-Ins2 remained unchanged in normoglycemic ob/ob mice but was reduced in islets of hyperglycemic db/db mice, as observed for circHIPK3 and ciRS-7 [40][46]. In addition, the level of human ci-INS was lower in islets of T2D donors and was inversely correlated with glycated hemoglobin (HbA1c) levels [46]. Taken together, these data suggest that ci-Ins2 might regulate insulin secretion in part through TDP-43 binding and contribute to β-cell dysfunction in the pathogenesis of T2D.
In addition to involving insulin secretion, circRNAs also impact β-cell apoptosis or proliferation. circAFF1 is exonic from the AF4/FMR2 family member 1 (AFF1) gene. The level of circAFF1 did not change in the type 1 or T2D model, but silencing circAFF1 enhanced β-cell apoptosis in vitro. Nevertheless, circAFF1 did not affect the capacities of proliferation, insulin content, or secretion in β-cells [40]. In contrast, circTulp4 promoted β-cell function by inducing the expression of the cholesterol esterification-related gene, sterol O-acyltransferase 1 (SOAT1), through inhibiting miR-7222-3p [48]. The accumulation of soat1 activated cyclin D1 expression and thus promoted cell cycle progression. Those findings suggested that circTulp4 might upregulate β-cell proliferation by the miR-7222-3p/soat1/cyclin D1 pathway [48].
Of note, more than 2600 circRNAs were recently identified in human pancreatic islets by CircleSeq [49]. One of the most abundant circRNAs in human islets, circCIRBP, demonstrated an association with the insulin secretory index in isolated human islets [49]. In addition, some islet circRNAs are expressed in peripheral blood, and circCAMSAP1 was correlated with T2D status [49].
3.2. circRNAs in IR
It is widely believed that IR is strongly associated with obesity and T2DM. Studies have proven the roles of miRNAs in regulating obesity and IR [50][51]. Until now, few comprehensive studies have specifically utilized the role of circRNAs in obesity-IR-T2DM settings. Nevertheless, a recent report of the involvement of circHIPK3 in hyperglycemia and IR by Cai et al. might open up this new world [52]. They demonstrated that increased circHIPK3 could sponge miR-192-5p, which targets and degrades Forkhead box protein O1 (FOXO1) mRNA, decreasing its expression in HepG2 cells [52]. FOXO1 is a transcription factor that plays vital roles in regulating gluconeogenesis and glycogenolysis by insulin signaling [53][54]. Moreover, FOXO1 inhibition increases uncoupling protein 1 (Ucp1) expression, which subsequently increases thermogenesis and reduces fat mass [55][56]. These findings suggest that circHIPK3 may increase IR and hyperglycemia by upregulating FOXO1. Even though those current studies did not directly link potential circRNA signatures and obesity-related IR, they highlight a future direction for further elucidation.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23031032
References
- Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 3 December 2020).
- Reaven, G.M. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 1988, 37, 1595–1607.
- Samuel, V.T.; Shulman, G.I. The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux. J. Clin. Investig. 2016, 126, 12–22.
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856.
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030.
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211.
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777.
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66.
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134.
- Taggart, A.J.; DeSimone, A.M.; Shih, J.S.; Filloux, M.E.; Fairbrother, W.G. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat. Struct. Mol. Biol. 2012, 19, 719–721.
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417.
- Zaphiropoulos, P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping. Proc. Natl. Acad. Sci. USA 1996, 93, 6536–6541.
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.L.; Yang, L.; Chen, L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624.
- Chen, W.; Schuman, E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016, 39, 597–604.
- Li, Y.; Zhao, J.; Yu, S.; Wang, Z.; He, X.; Su, Y.; Guo, T.; Sheng, H.; Chen, J.; Zheng, Q.; et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin. Chem. 2019, 65, 798–808.
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, 6357.
- Yu, L.; Gong, X.; Sun, L.; Zhou, Q.; Lu, B.; Zhu, L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE 2016, 11, e0158347.
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928.
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174, 350–362.e317.
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215.
- Yu, C.Y.; Li, T.C.; Wu, Y.Y.; Yeh, C.H.; Chiang, W.; Chuang, C.Y.; Kuo, H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017, 8, 1149.
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409.
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369.
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429.
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858.
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020, 12, 112.
- Zhao, Z.J.; Shen, J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017, 14, 514–521.
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435.
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594.
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010.
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29.
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475.
- Tian, Y.; Xu, J.; Du, X.; Fu, X. The interplay between noncoding RNAs and insulin in diabetes. Cancer Lett. 2018, 419, 53–63.
- Stoll, L.; Sobel, J.; Rodriguez-Trejo, A.; Guay, C.; Lee, K.; Veno, M.T.; Kjems, J.; Laybutt, D.R.; Regazzi, R. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metab. 2018, 9, 69–83.
- Motterle, A.; Gattesco, S.; Caille, D.; Meda, P.; Regazzi, R. Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 2015, 58, 1827–1835.
- Guay, C.; Jacovetti, C.; Nesca, V.; Motterle, A.; Tugay, K.; Regazzi, R. Emerging roles of non-coding RNAs in pancreatic beta-cell function and dysfunction. Diabetes Obes. Metab. 2012, 14 (Suppl. 3), 12–21.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Xu, H.; Guo, S.; Li, W.; Yu, P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015, 5, 12453.
- Latreille, M.; Hausser, J.; Stutzer, I.; Zhang, Q.; Hastoy, B.; Gargani, S.; Kerr-Conte, J.; Pattou, F.; Zavolan, M.; Esguerra, J.L.; et al. MicroRNA-7a regulates pancreatic beta cell function. J. Clin. Investig. 2014, 124, 2722–2735.
- Stoll, L.; Rodriguez-Trejo, A.; Guay, C.; Brozzi, F.; Bayazit, M.B.; Gattesco, S.; Menoud, V.; Sobel, J.; Marques, A.C.; Veno, M.T.; et al. A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 2020, 11, 5611.
- Araki, K.; Araki, A.; Honda, D.; Izumoto, T.; Hashizume, A.; Hijikata, Y.; Yamada, S.; Iguchi, Y.; Hara, A.; Ikumi, K.; et al. TDP-43 regulates early-phase insulin secretion via CaV1.2-mediated exocytosis in islets. J. Clin. Investig. 2019, 129, 3578–3593.
- Wu, L.; Xiong, L.; Li, J.; Peng, Z.; Zhang, L.; Shi, P.; Gong, Y.; Xiao, H. Circ-Tulp4 promotes beta-cell adaptation to lipotoxicity by regulating soat1 expression. J. Mol. Endocrinol. 2020, 65, 149–161.
- Haque, S.; Ames, R.M.; Moore, K.; Lee, B.P.; Jeffery, N.; Harries, L.W. Islet-expressed circular RNAs are associated with type 2 diabetes status in human primary islets and in peripheral blood. BMC Med. Genom. 2020, 13, 64.
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. miRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744.
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653.
- Cai, H.; Jiang, Z.; Yang, X.; Lin, J.; Cai, Q.; Li, X. Circular RNA HIPK3 contributes to hyperglycemia and insulin homeostasis by sponging miR-192-5p and upregulating transcription factor forkhead box O1. Endocr. J. 2020, 67, 397–408.
- Matsumoto, M.; Pocai, A.; Rossetti, L.; Depinho, R.A.; Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007, 6, 208–216.
- Wu, Y.; Pan, Q.; Yan, H.; Zhang, K.; Guo, X.; Xu, Z.; Yang, W.; Qi, Y.; Guo, C.A.; Hornsby, C.; et al. Novel Mechanism of Foxo1 Phosphorylation in Glucagon Signaling in Control of Glucose Homeostasis. Diabetes 2018, 67, 2167–2182.
- Nakae, J.; Cao, Y.; Oki, M.; Orba, Y.; Sawa, H.; Kiyonari, H.; Iskandar, K.; Suga, K.; Lombes, M.; Hayashi, Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 2008, 57, 563–576.
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11.
This entry is offline, you can click here to edit this entry!
