Liver diseases are one of the main causes of mortality. In this regard, the development of new ways of reparative processes stimulation is relevant. Macrophages play a leading role in the regulation of liver homeostasis in physiological conditions and in pathology. In this regard, the development of new liver treatment methods is impossible without taking into account this cell population. Resident macrophages of the liver, Kupffer cells, represent a unique cell population, first of all, due to their development. Most of the liver macrophages belong to the self-sustaining macrophage cell population, whose origin is not bone marrow. In addition, Kupffer cells are involved in such processes as regulation of hepatocyte proliferation and apoptosis, remodeling of the intercellular matrix, lipid metabolism, protective function, etc. Such a broad spectrum of liver macrophage functions indicates their high functional plasticity. The Recent data on the development, phenotypic and functional plasticity, and participation in the reparative processes of liver macrophages: resident macrophages (Kupffer cells) and bone marrow-derived macrophages were summarized.
- monocytes
- macrophages
- Kupffer cells
- liver
- regeneration
1. Introduction
2. Macrophage Population of the Liver
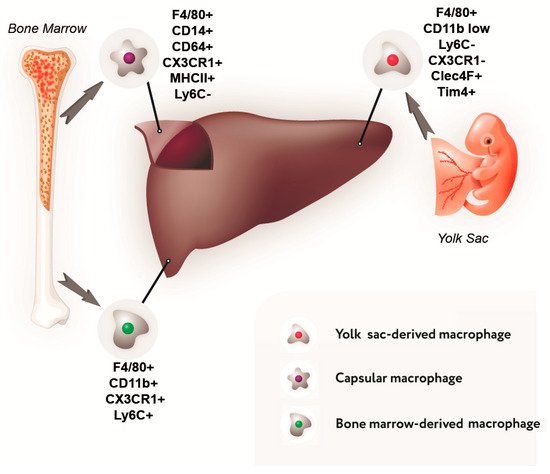
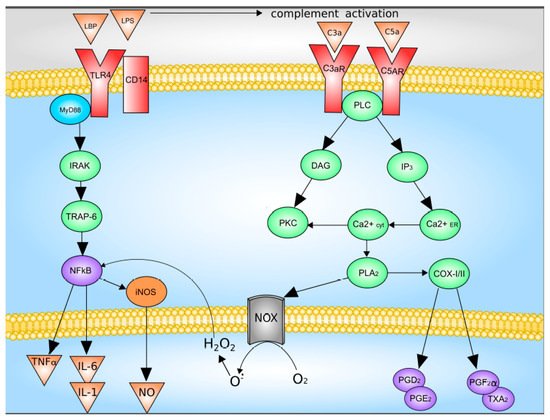
3. The Diversity of Macrophage Functional Types
This entry is adapted from the peer-reviewed paper 10.3390/cells8091032
References
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761.
- Ganne-Carrié, N. Epidemiology of liver cirrhosis. Rev. Prat. 2017, 67, 726–730.
- Nicolas, C.T.; Hickey, R.D.; Chen, H.S.; Mao, S.A.; Lopera Higuita, M.; Wang, Y.; Nyberg, S.L. Concise Review: Liver Regenerative Medicine: From Hepatocyte Transplantation to Bioartificial Livers and Bioengineered Grafts. Stem Cells 2017, 35, 42–50.
- Zhang, J.; Zhao, X.; Liang, L.; Li, J.; Demirci, U.; Wang, S. A decade of progress in liver regenerative medicine. Biomaterials 2018, 157, 161–176.
- Bilzer, M.; Roggel, F.; Gerbes, A.L. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006, 26, 1175–1186.
- Liaskou, E.; Wilson, D.V.; Oo, Y.H. Innate immune cells in liver inflammation. Mediators Inflamm. 2012, 2012, 949157.
- Dong, X.; Liu, J.; Xu, Y.; Cao, H. Role of macrophages in experimental liver injury and repair in mice. Exp. Ther. Med. 2019, 17, 3835–3847.
- Michalopoulos, G.K. Advances in liver regeneration. Expert Rev. Gastroenterol. Hepatol. 2014, 8, 897–907.
- Guillot, A.; Tacke, F. Liver Macrophages: Old Dogmas and New Insights. Hepatol. Commun. 2019, 3, 730–743.
- Wynn, T.A.; Barron, L. Macrophages: master regulators of inflammation and fibrosis. Semin. Liver Dis. 2010, 30, 245–257.
- Beljaars, L.; Schippers, M.; Reker-Smit, C.; Martinez, F.O.; Helming, L.; Poelstra, K.; Melgert, B.N. Hepatic Localization of Macrophage Phenotypes during Fibrogenesis and Resolution of Fibrosis in Mice and Humans. Front. Immunol. 2014, 5, 430.
- Tacke, F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017, 66, 1300–1312.
- Keirsse, J.; Van Damme, H.; Geeraerts, X.; Beschin, A.; Raes, G.; Van Ginderachter, J.A. The role of hepatic macrophages in liver metastasis. Cell. Immunol. 2018, 330, 202–215.
- Yeh, C.-C.; Chao, K.-C.; Huang, S.J. Innate Immunity, Decidual Cells, and Preeclampsia. Reprod. Sci. 2013, 20, 339–353.
- Chazaud, B. Macrophages: Supportive cells for tissue repair and regeneration. Immunobiology 2014, 219, 172–178.
- Nishiyama, K.; Nakashima, H.; Ikarashi, M.; Kinoshita, M.; Nakashima, M.; Aosasa, S.; Seki, S.; Yamamoto, J. Mouse CD11b+Kupffer cells recruited from bone marrow accelerate liver regeneration after partial hepatectomy. PLoS ONE 2015, 10, 1–16.
- Ikarashi, M.; Nakashima, H.; Kinoshita, M.; Sato, A.; Nakashima, M.; Miyazaki, H.; Nishiyama, K.; Yamamoto, J.; Seki, S. Distinct development and functions of resident and recruited liver Kupffer cells/macrophages. J. Leukoc. Biol. 2013, 94, 1325–1336.
- Zigmond, E.; Samia-Grinberg, S.; Pasmanik-Chor, M.; Brazowski, E.; Shibolet, O.; Halpern, Z.; Varol, C. Infiltrating Monocyte-Derived Macrophages and Resident Kupffer Cells Display Different Ontogeny and Functions in Acute Liver Injury. J. Immunol. 2014, 193, 344–353.
- Ju, C.; Tacke, F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell. Mol. Immunol. 2016, 13, 316–327.
- Perdiguero, E.G.; Geissmann, F. The development and maintenance of resident macrophages. Nat. Immunol. 2016, 17, 2–8.
- You, Q.; Holt, M.; Yin, H.; Li, G.; Hu, C.J.; Ju, C. Role of hepatic resident and infiltrating macrophages in liver repair after acute injury. Biochem. Pharmacol. 2013, 86, 836–843.
- Armbrust, T.; Ramadori, G. Functional characterization of two different Kupffer cell populations of normal rat liver. J. Hepatol. 1996, 25, 518–528.
- He, Y.; Sadahiro, T.; Noh, S.I.; Wang, H.; Todo, T.; Chai, N.; Klein, A.S.; Wu, G.D. Flow cytometric isolation and phenotypic characterization of two subsets of ED2+ (CD163) hepatic macrophages in rats. Hepatol. Res. 2009, 39, 1208–1218.
- Gottfried, E.; Kunz-Schughart, L.A.; Weber, A.; Rehli, M.; Peuker, A.; Müller, A.; Kastenberger, M.; Brockhoff, G.; Andreesen, R.; Kreutz, M. Expression of CD68 in Non-Myeloid Cell Types. Scand. J. Immunol. 2008, 67, 453–463.
- Schittenhelm, L.; Hilkens, C.M.; Morrison, V.L. β2 Integrins As Regulators of Dendritic Cell, Monocyte, and Macrophage Function. Front. Immunol. 2017, 8, 1866.
- Sanchez-Madrid, F.; Simon, P.; Thompson, S.; Springer, T.A. Mapping of antigenic and functional epitopes on the alpha- and beta-subunits of two related mouse glycoproteins involved in cell interactions, LFA-1 and Mac-1. J. Exp. Med. 1983, 158, 586–602.
- Kinoshita, M.; Uchida, T.; Sato, A.; Nakashima, M.; Nakashima, H.; Shono, S.; Habu, Y.; Miyazaki, H.; Hiroi, S.; Seki, S. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J. Hepatol. 2010, 53, 903–910.
- Hoeffel, G.; Ginhoux, F. Fetal monocytes and the origins of tissue-resident macrophages. Cell. Immunol. 2018, 330, 5–15.
- Movita, D.; Kreefft, K.; Biesta, P.; van Oudenaren, A.; Leenen, P.J.M.; Janssen, H.L.A.; Boonstra, A. Kupffer cells express a unique combination of phenotypic and functional characteristics compared with splenic and peritoneal macrophages. J. Leukoc. Biol. 2012, 92, 723–733.
- Haldar, M.; Murphy, K.M. Origin, development, and homeostasis of tissue-resident macrophages. Immunol. Rev. 2014, 262, 25–35.
- MacParland, S.A.; Liu, J.C.; Ma, X.Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9.
- Krenkel, O.; Hundertmark, J.; Abdallah, A.T.; Kohlhepp, M.; Puengel, T.; Roth, T.; Branco, D.P.P.; Mossanen, J.C.; Luedde, T.; Trautwein, C.; et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 2019.
- Graubardt, N.; Vugman, M.; Mouhadeb, O.; Caliari, G.; Pasmanik-Chor, M.; Reuveni, D.; Zigmond, E.; Brazowski, E.; David, E.; Chappell-Maor, L.; et al. Ly6Chimonocytes and their macrophage descendants regulate neutrophil function and clearance in acetaminophen-induced liver injury. Front. Immunol. 2017.
- Yin, S.; Wang, H.; Park, O.; Wei, W.; Shen, J.; Gao, B. Enhanced liver regeneration in IL-10-deficient mice after partial hepatectomy via stimulating inflammatory response and activating hepatocyte STAT3. Am. J. Pathol. 2011, 178, 1614–1621.
- Behnke, K.; Zhuang, Y.; Xu, H.C.; Sundaram, B.; Reich, M.; Shinde, P.V.; Huang, J.; Modares, N.F.; Tumanov, A.V.; Polz, R.; et al. B Cell-Mediated Maintenance of Cluster of Differentiation 169-Positive Cells Is Critical for Liver Regeneration. Hepatology 2018, 68, 2348–2361.
- González-Domínguez, É.; Samaniego, R.; Flores-Sevilla, J.L.; Campos-Campos, S.F.; Gómez-Campos, G.; Salas, A.; Campos-Peña, V.; Corbí, Á.L.; Sánchez-Mateos, P.; Sánchez-Torres, C. CD163L1 and CLEC5A discriminate subsets of human resident and inflammatory macrophages in vivo. J. Leukoc. Biol. 2015, 98, 453–466.
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20.
- Malyshev, I.; Malyshev, Y. Current Concept and Update of the Macrophage Plasticity Concept: Intracellular Mechanisms of Reprogramming and M3 Macrophage “Switch” Phenotype. Biomed Res. Int. 2015, 2015, 1–22.
- Ren, C.-X.; Leng, R.-X.; Fan, Y.-G.; Pan, H.-F.; Li, B.-Z.; Wu, C.-H.; Wu, Q.; Wang, N.-N.; Xiong, Q.-R.; Geng, X.-P.; et al. Intratumoral and peritumoral expression of CD68 and CD206 in hepatocellular carcinoma and their prognostic value. Oncol. Rep. 2017, 38, 886–898.
- Mehal, W.Z. The inflammasome in liver injury and non-alcoholic fatty liver disease. Dig. Dis. 2014, 32, 507–515.
- Wree, A.; Eguchi, A.; Mcgeough, M.D.; Pena, C.A.; Johnson, C.D.; Canbay, A.; Hoffman, H.M.; Feldstein, A.E. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology 2014, 59, 898–910.
- Murray, P.J.; Wynn, T.A. Obstacles and opportunities for understanding macrophage polarization. J. Leukoc. Biol. 2011, 89, 557–563.
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014, 6, 13.
- Okizaki, S.; Ito, Y.; Hosono, K.; Oba, K.; Ohkubo, H.; Amano, H.; Shichiri, M.; Majima, M. Suppressed recruitment of alternatively activated macrophages reduces TGF-β1 and impairs wound healing in streptozotocin-induced diabetic mice. Biomed. Pharmacother. 2015, 70, 317–325.
- Nakajima, H.; Uchida, K.; Guerrero, A.R.; Watanabe, S.; Sugita, D.; Takeura, N.; Yoshida, A.; Long, G.; Wright, K.T.; Johnson, W.E.B.; et al. Transplantation of Mesenchymal Stem Cells Promotes an Alternative Pathway of Macrophage Activation and Functional Recovery after Spinal Cord Injury. J. Neurotrauma 2012, 29, 1614–1625.
- Singla, D.K.; Singla, R.D.; Abdelli, L.S.; Glass, C. Fibroblast Growth Factor-9 Enhances M2 Macrophage Differentiation and Attenuates Adverse Cardiac Remodeling in the Infarcted Diabetic Heart. PLoS ONE 2015, 10, e0120739.
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566.
- Kiguchi, N.; Kobayashi, Y.; Saika, F.; Sakaguchi, H.; Maeda, T.; Kishioka, S. Peripheral interleukin-4 ameliorates inflammatory macrophage-dependent neuropathic pain. Pain 2015, 156, 684–693.
- Yang, S.-L.; Chen, S.-L.; Wu, J.-Y.; Ho, T.-C.; Tsao, Y.-P. Pigment epithelium-derived factor induces interleukin-10 expression in human macrophages by induction of PPAR gamma. Life Sci. 2010, 87, 26–35.
- Kole, A.; Kelsall, B.L.; Valatas, V.; He, J.; Rivollier, A. Inflammation switches the differentiation program of Ly6C hi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 2012, 209, 139–155.
- Bain, C.C.; Scott, C.L.; Uronen-Hansson, H.; Gudjonsson, S.; Jansson, O.; Grip, O.; Guilliams, M.; Malissen, B.; Agace, W.W.; Mowat, A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013, 6, 498–510.
- Yin, G.; Zhu, X.; Guo, C.; Yang, Y.; Han, T.; Chen, L.; Yin, W.; Gao, P.; Zhang, H.; Geng, J.; et al. Differential expression of estradiol and estrogen receptor α in severe preeclamptic pregnancies compared with normal pregnancies. Mol. Med. Rep. 2013, 7, 981–985.
- Fabriek, B.O.; van Bruggen, R.; Deng, D.M.; Ligtenberg, A.J.M.; Nazmi, K.; Schornagel, K.; Vloet, R.P.M.; Dijkstra, C.D.; van den Berg, T.K. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 2009, 113, 887–892.
- Martinez-Pomares, L. The mannose receptor. J. Leukoc. Biol. 2012, 92, 1177–1186.
- Stahl, P.D.; Ezekowitz, R.A. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. 1998, 10, 50–55.
- Lokhonina, A.; Elchaninov, A.; Fatkhudinov, T.; Makarov, A.; Arutyunyan, I.; Grinberg, M.; Glinkina, V.; Surovtsev, V.; Bolshakova, G.; Goldshtein, D.; et al. Activated Macrophages of Monocytic Origin Predominantly Express Proinflammatory Cytokine Genes, Whereas Kupffer Cells Predominantly Express Anti-Inflammatory Cytokine Genes. Biomed Res. Int. 2019, 2019, 1–13.
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084.
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; de Dios Ruiz-Rosado, J.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-de-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342.
