RNA silencing is as an adaptive immune response in plants that limits the accumulation or spread of invading viruses. Successful virus infection entails countering the RNA silencing machinery for efficient replication and systemic spread in the host. Tungro is a viral disease caused by a complex of two viruses. The Rice tungro bacilliform virus (RTBV) is the major determinant of the disease while Rice tungro spherical virus (RTSV) accentuates the symptoms. The RTBV ORF-IV is a weak suppressor of host silencing, but its suppression activity is augmented in the presence of RTSV protease, and coat protein 3. RTSV components might also have a possible role in enhancing the suppression of the cell-to-cell spread of silencing.
1. Introduction
The small RNA mediated gene silencing is a sequence-specific regulatory phenomenon resulting in mRNA degradation or translation inhibition at the post-transcriptional level and DNA methylation at the transcriptional level. The process involves generation of 21 to 24 nt long double stranded RNA intermediates by the action of specific Dicer like (DCL) molecules that guide the silencing through Argonaute (AGO) containing protein complexes. Silencing works as an effective tool to counter virus infection.
For successfully invading the hosts, viruses have co-evolved tools to effectively suppress the components of host silencing. The suppression activity has evolved independently such that existing viral proteins performing certain vital functions have gained an additional role of suppressing RNA silencing. The viral encoded suppressor molecules have been recognized as pathogenicity determinants as they have the ability to counteract antiviral silencing and play an important role in virulence. The suppressor proteins do not share any co-evolutionary patterns and they have unique functional domains and properties, which makes their identification difficult. Each suppressor may have distinctive mechanism of interacting with the components of host RNA silencing machinery. A number of in planta assays are available to screen the suppressor activity based on the principle of suppression of the RNA silencing of a reporter transgene, that results in expression of the reporter gene.
Tungro disease of rice is considered to be one of the most damaging viral diseases of rice prevalent in South and Southeast Asia and accounts for huge economic loss. Stunting, yellow orange discoloration of leaves and twisting of the leaf tips are the major symptoms in plants infected with virus. It is caused by a complex of two viruses, transmitted together by the green leafhoppers (GLH)—Nephotettix virescens and N. nigropictus. The Rice tungro spherical virus (RTSV), a positive single strand RNA virus in the Secoviridae family, is required for transmission of the disease. Single infection by RTSV does not produce any detectable symptoms of the disease. Its genome consists of 12433-nt RNA with a poly-A tail at the 3′ end that codes for a large polyprotein, which is cleaved to form three mature capsid proteins and other viral proteins. Rice tungro bacilliform virus (RTBV), a double strand DNA pararetrovirus virus in the Caulimoviridae family, is considered to be the major cause for the manifestation of the disease. The RTBV genome contains four open reading frames (ORFs). The functions of ORF-I and ORF-II are yet not defined. The ORF-III codes for a large polyprotein, which is processed in to a movement protein (MP), a coat protein (CP), an aspartate protease (AP), and replicase proteins. The ORF-IV transcript undergoes splicing to generate a protein that has a sequence motif similar to leucine zipper, although it lacks the DNA binding region. Though RTBV is the main determinant of disease symptoms, the symptoms get accentuated in the presence of RTSV. The results obtained in our work suggest that the complexity of the disease is likely driven by the combined suppressor action of the two viral components of the tungro system.
2. ORF-IV Can Suppress Pre-Established RNA Silencing
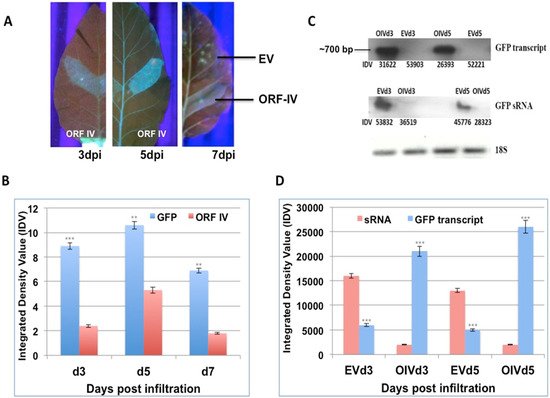
Leaves of stably silent GFP tobacco lines were infiltrated on one side of the midrib with Agrobacterium cultures containing the individual ORFs of RTBV (ORF-I, ORF-II, and ORF-IV) and RTSV (protease, polymerase, and coat protein 3). Leaves were collected at 3, 5, and 7 days post infiltration (dpi) and scanned under UV-light to observe for GFP fluorescence, as proof for the reversal of silencing by the infiltrated suppressor. The RTBV ORF-I, ORF-II, and the RTSV ORFs did not show clear suppressor activity in individual infiltrations. In regions infiltrated with RTBV ORF-IV, a low level of fluorescence was observed in leaves at 3 dpi, while significant fluorescence could be observed at 5 dpi, and this dipped again by 7 dpi (Figure 1A). No fluorescence was detected in the regions infiltrated with empty vector that served as the control. This showed that ORF-IV acts as a weak suppressor of the S-PTGS or pre-established stable silencing.
Figure 1. Reversal of GFP silencing by RTBV ORF-IV and its molecular analysis. (A) The different panels exhibit GFP fluorescence in the region infiltrated with ORF-IV at 3, 5, and 7 days post infiltration (dpi). The region infiltrated with the empty vector (EV) served as the control. (B) The graph represents the normalized Integrated Density Values (IDV) for the GFP and ORF-IV transcripts relative to 18S control cDNA at 3 dpi (d3), 5 dpi (d5), and 7 dpi (d7). (C) Northern blots of GFP transcript and GFP sRNA in regions infiltrated with ORF-IV (OIV) and EV. The 18S cDNA band served as the control. (D) The graph represents the normalized values of GFP transcripts and GFP siRNA as measured in the infiltrated regions at 3 dpi and 5 dpi. **, *** Significance at probability levels of 1% and 0.1%, respectively (ANOVA single factor).
The suppressor activity was confirmed by validating the levels of the GFP transcript and GFP small RNAs using northern blot (Figure 1B). It was seen that GFP transcripts accumulated only in the regions where ORF-IV was transcribed, indicating its interference with the silencing machinery. The time kinetics revealed that the GFP transcript started forming by 3 dpi, even though significant florescence levels could be detected at 5 dpi (Figure 1C). Correspondingly, low levels of GFP small RNAs were observed in leaf regions infiltrated with ORF-IV, while prominent levels of the GFP small RNAs were detected in the empty vector infiltrated leaf sections (Figure 1B,D). The negative correlation between the small RNA and ORF-IV transcript accumulation confirmed the suppressor action of ORF-IV.
3. RTSV Coat Protein Enhances the Suppressor Activity of ORF-IV
To further investigate the nature of the RNA silencing suppression activity of RTBV ORF-IV in the presence of RTSV, the same assay system was employed. The leaves were infiltrated, respectively, with ORF-I and ORF-II of RTBV and RTSV ORF encoding protease, coat protein 3 (CP3), and polymerase as co-infiltrations with the ORF-IV (Figure 2). The infiltrations with ORF-IV alone served as the reference point for assaying the suppressor activity.
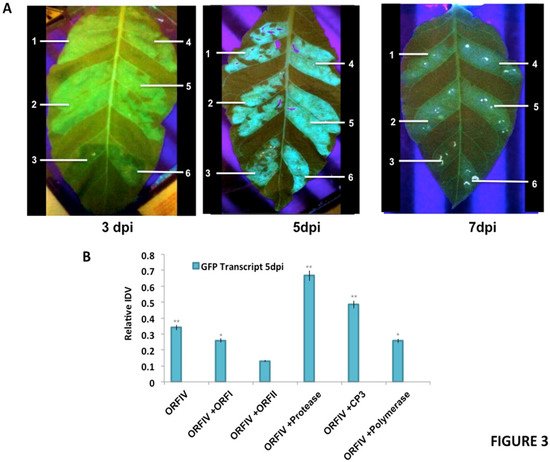
Figure 2. Reversal of GFP silencing by co-infiltration of RTBV and RTSV ORFs. (A) The different panels exhibit representative pictures to show GFP fluorescence in the infiltrated regions at 3, 5, and 7 days post infiltration (dpi). The markings indicate the region infiltrated with (1) ORF-IV construct, (2) ORF-IV co-infiltrated with RTBV-ORF-I, (3) ORF-IV co-infiltrated with RTBV ORF-II, (4) ORF-IV co-infiltrated with RTSV ORF coding for coat protein 3, (5) ORF-IV co-infiltrated with RTSV ORF coding for protease, and (6) ORF-IV co-infiltrated with RTSV ORF coding for polymerase. (B) Normalized values for GFP transcripts in the different infiltrated regions at 5 dpi, confirmed using RT PCR. *, ** Significance at probability levels of 5% and 1%, respectively (ANOVA single factor).
It was observed that at 3 dpi, enhanced suppressor activity is observed in regions co-infiltrated with ORF-IV and RTSV ORF, encoding CP3 and polymerase. At 5 dpi, the suppression activity was significantly enhanced in individual co-infiltrations with RTSV ORFs, but decreased in the presence of the RTBV ORF-II. The enhanced suppression activities in the regions co-infiltrated with RTBV ORF-IV and RTSV ORFs encoding protease as well as CP3 were sustained up to 7 dpi (Figure 2A).
To validate the results, molecular analysis of the co-infiltrated zones was performed to check for the presence of GFP as well as the ORF transcripts. For each transcript, the relative band intensity values were calculated by normalizing with respect to the 18S control. Normalized values for GFP transcripts with respect to ORFIV in the different infiltrated regions at 5 dpi were plotted (Figure 2B). A higher level of expression was seen for GFP in the regions where ORF-IV was co-infiltrated with RTSV ORFs coding for protease and CP3, indicating their role in the enhancement of the suppression activity encoded by RTBV. This clearly demonstrates that the early onset of suppression of ORF-IV is enhanced and sustained by the activity of RTSV ORFs, which may potentially lead to a potent infection in the rice plant. It was also observed that, at 3 dpi, the respective presence of RTSV protease and CP3 caused a transient spread of silencing beyond the infiltrated zone (Figure 2A). This indicates the suppression of both the local siRNAs as well as the mobile signals responsible for the systemic spread of silencing; however, further investigation is required to understand the mechanism of action.
4. RTBV ORF-IV Interacts with RTSV CP3 Protein
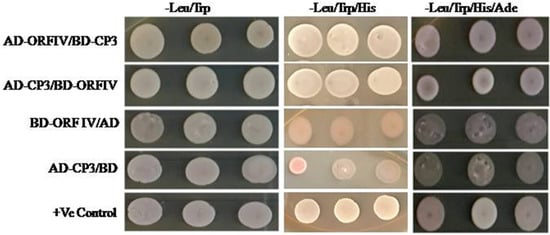
To confirm if enhancement in ORF-IV suppressor activity by RTSV proteins involves their direct interaction, yeast two hybrid (Y2H) assay was performed. Yeast cells co-expressing binding domain (BD) fused with ORF-IV and activation domain (AD) fused with CP3 could grow on both three-drop-out media (without amino acids Leu: leucine, TRP: tryptophan, and His: histidine) and four-drop-out media (without amino acids: Leu, TRP, His, and Ade: adenine). The results show a direct and strong interaction of RTBV ORF-IV with RTSV CP3 (Figure 3). The other proteins of RTSV coding for CP1, CP2, P1, polymerase, and protease did not show any interaction with RTBV ORF-IV. The observation indicated that the two proteins might be suppressing the silencing pathway by either targeting different components or acting independently on a common intermediate of the silencing pathway. Further work will, therefore, be required to unravel, the nature of protein interactions in vivo and their association in suppressing the RNAi driven host defense.
Figure 3. Yeast two-hybrid assay for direct interaction study. Yeast colonies of pGAD-CP3 and pGBD-ORF-IV were co-transformed and selected on (i) two drop out plate (Leu− Trp−), (ii) triple drop out (Leu− Trp− His−), and (iii) triple drop out (Leu− Trp− His−) supplemented with 5 μ M 3-AT. Co-infiltration of pGAD-CP3 with pGBD and pGAD with pGBD-ORF-IV was performed as the control.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms10020197