Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Hyperspectral remote sensing provides image data with very high spectral resolution. This high resolution allows subtle differences in plant health to be recognized. Such a multidimensional data space, generated by hyperspectral sensors, has given rise to new approaches and methods for analyzing hyperspectral data.
- remote sensing
- hyperspectral
- plant diseases
- early detection
- oil palm
- citrus
- cereals
- solanaceae
1. Introduction
The spread of various, including invasive, plant diseases and pests is one of the most important problems in modern agriculture [1]. Therefore, to solve these relevant problems, the timely monitoring of plant diseases and pests is necessary. Remote sensing methods hold great promise for solving these problems [2]. Remote sensing data can identify crop conditions, including diseases, and provide useful information for specific agricultural management practices [3,4].
There are two types of remote sensing technologies: passive (such as optical) and active remote sensing (such as LiDAR and radar). Passive optical remote sensing is usually divided into two groups based on the spectral resolution of the sensors used: multispectral and hyperspectral remote sensing [5]. Hyperspectral sensing shows great potential as a non-invasive and non-destructive tool for monitoring biotic and abiotic plant stress among passive remote sensing methods, which measure reflected solar radiation [6]. This method collects and stores information from the spectroscopy of an object in a spectral cube that contains spatial information and hundreds of contiguous wavelengths in the third dimension. Hyperspectral imaging offers many opportunities for the early recognition of plant diseases by providing preliminary indicators through subtle changes in spectral reflectance due to absorption or reflection. Hyperspectral images with hundreds of spectral bands can provide detailed spectral portraits, hence, they are better able to detect subtle variations in soil, canopies or individual leaves. Thus, hyperspectral images can be used to solve a wider class of problems for the accurate and timely determination of the physiological status of agricultural crops. The early identification of disease spread and pest outbreaks may avoid not only significant crop loss, but also reduce pesticides usage and mitigate their negative impacts on human health and the environment, thus, improving the existing IPM [7,8].
In recent years, a wide range of miniature hyperspectral sensors available for commercial use have been developed, such as Micro- and Nano-Hyperspec (Headwall Photonics Inc., Boston, MA, USA), HySpex VNIR (HySpex, Skedsmo, Skjetten, Norway) and FireflEYE (Cubert GmbH, Ulm, Germany) [9]. These sensors can be installed on manned or unmanned airborne platforms (for example, airplanes, helicopters, and UAVs) to obtain hyperspectral imaging and support various monitoring missions [10,11].
There are various types of hyperspectral cameras, e.g., push-broom cameras, whisk-broom cameras and snapshot cameras. The measurement principle of each sensor type depends on its ability to obtain the whole picture (snapshot) at one time, one line of the picture (push broom) or one point of the picture (whisk broom) [12].
The general routine of collecting and processing hyperspectral images is presented in Figure 1. The light reflected from plant leaves is collected by the hyperspectral camera (Figure 1A) [13]. A hyperspectral data cube (Figure 1B) is obtained from the hyperspectral camera. Then various data normalization (Figure 1C) and feature extraction (Figure 1D) algorithms are applied to reduce the data’s dimensionality. Finally, different automatization techniques are used to automate the classification process (Figure 1E).
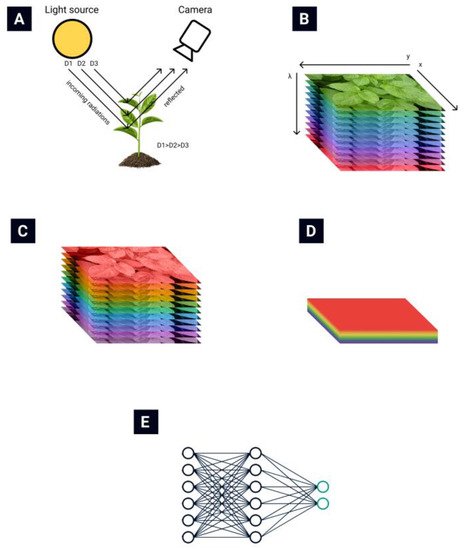
Hyperspectral remote sensing provides image data with very high spectral resolution [16,17]. This high resolution allows subtle differences in plant health to be recognized. Such a multidimensional data space, generated by hyperspectral sensors, has given rise to new approaches and methods for analyzing hyperspectral data [18,19].
For a long time, feature extraction methods have been used that reduce the data dimension without loss (or with minimal loss) of the original information on which the classification of hyperspectral images is based [20]. One of the most widely used dimensionality reduction techniques in HRS is principal component analysis (PCA). PCA computes orthogonal projections that maximize data variance and outputs the dataset in a new, uncorrelated coordinate system. Unfortunately, the informational content of hyperspectral images does not always coincide with such projections [21]. Thus, other methods are also used for feature extraction. The common methods for extracting hyperspectral data used in pathological research traditionally include PCA [22], derivative analysis [23], wavelet methods and correlation plots [24]. Alternatively, the hyperspectral image data can be processed at the image level to extract either spatial representation alone or joint spatial spectral information. If only spatial features are considered, for example, when studying structural and morphological features, spatial patterns among neighboring pixels with relation to the current pixel in the hyperspectral image will be extracted. Machine vision techniques, such as using a two-dimensional CNN, with a p × p chunk of input pixel data have been implemented to automatically generate high-level spatial structures. Extraction of spatial characteristics, in tandem with spectral elements, has been shown to significantly improve model performance. [25]. The use of spatial spectral characteristics can be achieved using two approaches: (i) by separately extracting spatial characteristics using CNN [26,27] and combining data from a spectral extractor using RNN, or LSTM [27,28]; and (ii) by using three-dimensional patterns in hyperspectral data cubes (p × p × b) associated with p × p spatially adjacent pixels and b spectral bands to take full advantage of important distinctive patterns.
2. Hyperspectral Remote Sensing for Early Plant Disease Detection
It was believed that, due to the lack of interaction between specialists in engineering and biology, there is a significant gap in the scientific basis for planning an experiment to use remote sensing data in determining plant state. Although the review above demonstrates the practical possibility of late and early detection of plant diseases using HRS, it also reveals differences in the technical results (range of important bands) between researchers, which indicates an insufficient study of the experimental methodology, as can be seen from Table 1, Table 2, Table 3 and Table 4.
As a result of hyperspectral remote sensing, for each pixel of a scene, we get a random vector, which can be considered the result of a random experiment. The outcome of a random experiment can be favorable or unfavorable, which is associated with the detection or non-detection of a disease in the space reflected by a particular pixel. Accordingly, these vectors can be processed by methods developed in the theory of probability and in mathematical statistics, which make it possible to effectively determine the characteristics of a random experiment. In this case, the tasks of data normalization and the allocation of those frequency bands (important bands) that make the greatest contribution to the outcomes of experiments (favorable or unfavorable) and, accordingly, are the most informative for identifying diseases, can be solved. The selection of important bands is a critical step in the detection of plant diseases using HRS. As a rule, data normalization is carried out first to get rid of noise. Then, various algorithms are applied to identify important bands, such as Savitzky–Golay filtering [50,51,58,81,82,83,99,100,109,131,132,147]; the Mann–Whitney U test [52,54]; coefficient of variation [60]; PCA [74,76,79,92,95,99,109,133]; SPA [78,102,106,107,108,144]; GA and BRT [106]; SAM [112,113,129,146].
The listed algorithms make it possible to achieve the determination of important bands. Various methods of machine learning allow achieving a fairly high accuracy in identifying diseases (between 60 and 95% accuracy) based on those data. However, from Table 1, Table 2, Table 3 and Table 4, we can conclude that even under very similar experimental conditions—For example when studying oil palms—Different sets of important bands are obtained at the output, often with a spread of more than 100 nm [52,53,54,55,56,57,58,59,60]. Xie et al., in [103], used five different algorithms to select important bands, taken from five different studies: t-test [164], Kullback–Leibler divergence [165], Chernoff bound [166], receiver operating characteristics [167] and the Wilcoxon test [168]. It is noteworthy that, in 4 tests out of 5, only 1 frequency out of 15 matched closely. In this case, the scatter of the ranges of all initially selected important bands was in the range from 400 to 850 nm, (400, 402, 403, 411, 413, 418, 419, 420, 422, 473, 642, 690, 722, 756 and 850 nm), i.e., practically in the entire range of the used sensor (380–1020 nm).
It was assume that, in the experiments on the same section of a field, repeated in different years or seasons, different important bands will likely be allocated when using automatic selection methods. Unfortunately, at the moment it is not possible to test this theory, since there are very few articles in which such experiments would be described.
Summarizing the topic of choosing the important bands for plant disease detection, we assume that it would be logical to focus on studying the bands of biochemical changes occurring in diseased plants and screening out the bands not related to the given disease, rather than using machine learning.
To successfully conduct the biological component of experiments on the HRS of plant diseases, it is necessary to understand that plant diseases are a particular case of plant stress. Plant diseases are processes that occur in plants under the influences of various reasons and which lead to their oppression and decreased productivity. Plant diseases are divided into two main groups: infectious and non-infectious [29,30]. The infectious plant diseases are caused by microorganisms (mainly fungi, bacteria, viruses and nematodes) or parasitic plants. The non-infectious diseases can be caused by genetic disorders or physiological metabolic disorders resulting from unfavorable environmental conditions [29,30]. Plant diseases almost always have visible symptoms that we can observe in a certain spectral range. In their early stages, such symptoms appear in the form of various chloroses or, less often, necrosis or pustules, with a huge variety of manifestations [169,170]. In the case of an asymptomatic course of the disease in its early stages, for example barley Ramularia disease caused by Ramularia collo-cygni [171], Fusarium head blight of different cereals caused by Fusarium culmorum [133] or soybean Sudden death caused by Fusarium virguliforme [172], early detection by remote sensing can be challenging.
Plant stress is a state of the plant in which it is influenced by unfavorable abiotic (light, heat, air, humidity, soil composition and relief conditions) and biotic factors (phytogenic, zoogenic, microbogenic and mycogenic). Plant responses to both abiotic and biotic stress is usually complex and includes both nonspecific (common for different stressors) and specific components. In a state of stress plants stop their growth, sharply reduce the activity of their root systems and reduce the intensity of photosynthesis and protein synthesis [173,174,175]. In a significant number of stressful situations, an immune response causes an increase of certain metabolites content, such as jasmonates or salicylates [175,176,177,178,179,180]. These reactions can be detected using hyperspectral sensors [181,182,183,184,185,186,187,188]. The study of plant stress using hyperspectral sensors is presented in a number of works [189,190,191], including those comparing the spectral portraits of plants simultaneously exposed to biotic and abiotic stress [192,193,194,195]. It is necessary to take into account many abiotic factors in addition to the possible influence of pathogens to accurately determine the reasons for stress manifestation [59,60,63,73,78,92,98,112,113,124,126,127,133,141]. Our analysis indicates that there is no unified methodology for conducting hyperspectral studies of plant diseases that takes into account the influence of abiotic factors. That is why we believe it is best to carry out experiments in laboratory conditions or in industrial greenhouses in order to partially or completely eliminate abiotic factors. Attempts to create various mobile vehicles operating at ground level whose purpose is to replace natural light sources with artificial light when using hyperspectral sensors in field experiments are described in [73,74,75,92,94,123]. This solves one of the main problems associated with the inhomogeneity of the solar spectrum due to changing weather conditions. Nevertheless, this approach cannot completely solve the problem of the influence of abiotic factors.
It would also be interesting to continue studies describing the definition of the phenotype and/or genotype of a plant and its influence on changes in the spectral portrait thereof [196,197,198,199,200,201]. Several studies reviewed describe that the host plant genotype has a significant impact on spectral reflectance and on the biochemical and physiological traits of the plants undergoing pathogen infection [76,78,110,111,112,113,124,126,127,140,141,147,148]. Therefore, it is very important to indicate the culture and cultivar of the studied plants. The exact indication of pathogens used for inoculation is also very important. We believe that comparisons of the spectral portraits of plants of different cultivars of the same crop is a primary task in creating a general methodology for detecting plant diseases using hyperspectral sensors. It is possible that the influence of chlorophyll fluorescence on the spectral portraits of plants and their related SVI may be a significant contribution to the solution of this problem [155,202,203,204,205]. Success in this area may allow the creation of patterns for determining phenotypes and plant cultivars within one crop, which will become the basis for a database of hyperspectral portraits of plants.
If we can confidently detect different types of plant stresses and distinguish plants infected with pathogens from healthy one and/or those affected by abiotic stresses, we can study the influence of the genotypic characteristics of a pathogen on the spectral profile of an infected plant. To do this, it is necessary to identify the differences between plants of the same phenotype as affected by pathogens with different genotypes. Since, for many pathogens, primarily micromycetes, the intrageneric and even intraspecific diversity is extremely high, it is necessary to investigate the possible differences in the spectral manifestations of symptoms, for example, between different species of fungi of the genus Fusarium or between different races of the brown rust pathogen (Puccinia triticina). The aim of such experiment will be to study the effect of the phenotypic and genotypic diversity of pathogens on the variability of spectral portraits of host plants. The visual manifestations of symptoms of yellow rust (Puccinia striiformis) caused by different races or different strains of Fusarium graminearum are often very similar. In the early stages of the disease, chlorosis caused by pathogens of different species may have similar spectral portraits, which become more distinguishable in the later stages of the disease, and, thus, is also an important direction for research [91,92,96,97,102,103,110,111,112,125,126,127,131,132]. The influence of plant resistance on the symptomatology of pathogenesis and works describing the difference in the data obtained in such cases is also worth mentioning [110,111,112,113,126,127,132,140,141,144,145,146,148]. The determination of resistant cultivars using hyperspectral sensing is also a promising area of research with great applied potential [126].
One more direction, which is important for the early detection of plant diseases using HRS, is the study of spectral portraits of pathogens themselves. Unfortunately, this is only possible for a small number of diseases, such as wheat powdery mildew caused by Blumeria graminis and wheat yellow rust of wheat caused by Puccinia striiformis, which show characteristic external symptoms in the early stages. Usually, these are diseases of fungal origin, where the object of detection is micromycete mycelium or spores on the leaf surface of a diseased plant. Disease detection by this method is considered in the example of wheat yellow rust, using pure fungal spore spectra as reference [147].
Pest control is also an important aspect of plant protection. We hypothesize that HRS can also be used to early detect such dangerous pests as the Colorado potato beetle (Leptinotarsa decemlineata), sunn pest (Eurygaster integriceps) [206], or western corn rootworm (Diabrotica virgifera virgifera), using spectral portraits of imago and different ages of larvae. Currently, a small number of works have been published on this topic [191,206,207,208,209,210], but we consider this direction to be very promising, especially for use in industrial greenhouses. Another possible direction of research is the detection of local outbreaks of pests outside farmlands, for example, locusts (Acridoidea) or beet webworms (Loxostege sticticalis), in order to eliminate them early before these pests can cause damage to yields.
It was believed that the effect of biochemical changes in plant tissues is critical for the early detection of plant diseases using passive sensors. The reflectance of light from plants leaves is dependent on multiple biophysical and biochemical interactions. The VIS range (400–700 nm) is influenced by pigment content. The NIR range (700–1100 nm) is influenced by leaf structure, internal scattering processes and by the light absorption by leaf water. The SWIR range (1100–2500) is influenced by chemicals and water composition [196,211,212,213,214,215,216].
The most investigated areas in this topic are the determination of changes in the content of water, nitrogen (N) in plants, as well as of chlorophyll or carotenoids, using various SVIs, which can be used to detect plant diseases. These techniques can be used to determine the nitrogen content of plants [217,218,219] and to detect plant stresses and diseases [56,57,78,220,221,222], including the early detection of plant diseases and pest infestations [147,154,156,157,223].
The topic of detecting individual chemical elements or chemical compounds, including volatiles, in plants is a less studied problem. In plant physiology, such elements are of great importance, such as nitrogen (N), one of the key components for chlorophyll; phosphorus (in the monovalent orthophosphate form H2PO4−), a key macronutrient; potassium (K+), influencing leaf color; calcium (Ca2+), which plays a fundamental physiological role in leaf structure and signaling; magnesium (Mg2+), an essential macronutrient for photosynthesis (as it is the central atom of chlorophyll); sulfur (S), in the form of sulfate; iron (Fe2+ or Fe3+), copper (Cu2+), manganese (Mn2+) and zinc (Zn2+), which are essential elements for plant growth and components of many enzymes; and the ions responsible for salination: Na+, K+, Ca2+, Mg2+ and Cl− [216]. The detection of these elements by HRS can be a key factor for identifying plant diseases at an early stage, since plant diseases are accompanied by a deficiency of some of the listed elements, which is the cause of chlorotic and necrotic changes in plant tissues [216]. Unfortunately, this task is difficult and poorly studied, but the following works prove the possibility of determining the chemical composition of plants in the VIS, NIR and SWIR ranges. Pandey et al. detected a wide range of macronutrients, namely N, P, K, Mg, Ca and S, and micronutrients, namely Fe, Mn, Cu and Zn, in maize and soybean plants [224]. Zhou et al. detected cadmium (Cd) concentrations in brown rice before harvest [225]. Ge et al. tried to analyze chlorophyll content (CHL), leaf water content (LWC), specific leaf area (SLA), nitrogen (N), phosphorus (P) and potassium (K) in maize using different SVIs but succeeded only with CHL and N [226]. Hu et al. proved to determine the content of Ca, Mg, Mo and Zn in wheat kernels [227].
The most difficult and interesting direction is the detection of the content of not individual elements, but more complex chemical compounds using HRS. As an example of such works, one can cite the articles by Gold et al., where the mechanisms of physiological changes in potato plants were considered when inoculated by Alternaria solani and Phytophthora infestans pathogens in the analytical example of the contents of foliar nitrogen, total phenolics, sugar and starch [112,113]. Fuentes et al. monitored the chemical fingerprints of different leaf samples and studied the correlation of aphid numbers in wheat plants with the presence and quantity alcohol, methane, hydrogen peroxide, aromatic compounds and amide functional groups compounds [228]. The paper [228] presented results on the implementation of SWIR HRS (1596–2396 nm) and a low-cost electronic nose (e-nose) coupled with machine learning. The authors believe that such study of plant physiology models open their use to assessing models of other biotic and abiotic stress effects on plants. Thus, the search for plant diseases at early stages using passive sensors, including hyperspectral ones, should be carried out in three main directions: the search for the characteristic immune response of the host plant to the pathogen, the search for characteristic symptoms of plant damage by the pathogen or the search for spectral portraits of the pathogen or pest itself. It is always necessary to take into account other stress factors affecting the spectral portrait of a diseased plant, which will allow us to accurately determine plant diseases using passive remote sensing.
Further development of experiment planning should be considered, preferably using a common methodology, so that there is an opportunity to adequately compare the results. An experiment tree, which will consider the physiological parameters of the plant should be designed [229]. All phases of the experiment should be considered and planned in advance, on the basis of the science of experiment planning, which is sufficiently well developed for applied physical research, based on the methods of probability theory and mathematical statistics. The following research phases for each type of sensors should be developed: laboratory research in deterministic conditions of deterministic parameters; the allocation of spectral bands responsible for certain parameters of plants (including diseases) in laboratory experiments; repetition (possibly multiple) of a laboratory experiment to collect statistics and validate; transfer of the experiment to field conditions to verify the correctness of the selected spectral bands. Such planning of experiments and the creation of a methodology for conducting them fills in the gaps associated with the lack of consideration of such factors as: different phenotypes of plants and their different spectral responses; various diseases and also their different spectral responses; the need to create and take into account a model of light propagation from an irradiating source to normalize hyperspectral imagery data [229,230,231,232].
This entry is adapted from the peer-reviewed paper 10.3390/s22030757
This entry is offline, you can click here to edit this entry!
