Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Veterinary Sciences
Porcine deltacoronavirus (PDCoV) is an emerging enteropathogenic coronavirus of swine that causes acute diarrhoea, vomiting, dehydration and mortality in seronegative neonatal piglets. PDCoV was first reported in Hong Kong in 2012 and its etiological features were first characterized in the United States in 2014. Currently, PDCoV is a concern due to its broad host range, including humans. Chickens, turkey poults, and gnotobiotic calves can be experimentally infected by PDCoV.
- porcine deltacoronaviruses
- origin
- evolution
- cross-species transmission
- zoonosis
1. Introduction
Coronaviruses (CoVs) are single-stranded, positive-sense RNA viruses belonging to the subfamily Orthocoronavirinae within the family of Coronaviridae. They are classified into four genera: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV). Coronaviruses are important pathogens of both humans and animals, causing diverse diseases. α-CoVs and β-CoVs infect mammals, while γ-CoVs and δ-CoVs primarily infect birds with some mammalian spill over [1][2][3]. Deltacoronaviruses were detected from the rectal swabs of mammalian species, including Asian leopard cats (Prionailurus bengalensis) and Chinese ferret badgers (Melogale moschata) in wet markets, during virological surveillance in southern China between 2005 and 2006 [4]. Since then, δ-CoVs have been detected from a wide range of birds and domestic pigs [5][6][7][8]. The Deltacoronavirus genus was defined by genomic sequence analysis of both avian and mammalian isolates [7]. The prototype porcine deltacoronavirus (PDCoV) HKU15 strain was reported in Hong Kong in 2012 [7]. However, its pathogenic potential was not recognized until 2014 when it was found to be the cause of pig diarrheic outbreaks, initially on several farms in Ohio, then spreading throughout the United States (US) and globally [9][10]. PDCoV infects the intestinal epithelial cells and causes acute watery diarrhoea, vomiting and dehydration in gnotobiotic and conventional piglets [9][11][12]. PDCoV causes clinical signs indistinguishable from those caused by other porcine enteric CoVs, such as porcine epidemic diarrhoea virus (PEDV) and transmissible gastroenteritis virus (TGEV) [13]. The clinical impact, prevalence, and disease severity of PDCoV in the field are lower than those of PEDV [14]. However, PDCoV has a broader host range and, unlike PEDV and TGEV, it could/can infect multiple species, including pigs, chickens, turkeys, cattle and humans [7][15][16][17][18]. This review focuses on the current knowledge on the origin, evolution, cross-species transmission and zoonotic potential of PDCoV. Additionally, we discuss the potential mechanisms for PDCoV interspecies transmission from birds to pigs, then to humans to help understand common mechanisms for the emergence of novel human CoVs, such as the highly pathogenic severe acute respiratory syndrome CoV (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
2. Origin of PDCoV
The Deltacoronavirus genus was first established by The International Committee on Taxonomy of Viruses (ICTV) in 2012 after the identification of three novel avian coronaviruses (bulbul CoV HKU11, thrush CoV HKU12, and munia CoV HKU13) [6] and seven other novel δ-CoVs (porcine CoV HKU15, white-eye CoV HKU16, sparrow CoV HKU17, magpie robin CoV HKU18, night heron CoV HKU19, wigeon CoV HKU20, and common moorhen CoV HKU21) of birds or pigs [7]. Interestingly, the replicase polyprotein gene (partial cds) of PDCoV HKU15 is closely related (nucleotide identity ≥ 98.88%) to those δ-CoVs (GenBank accession no. EF584909–EF584912) detected in 2006 from the Chinese ferret badgers at wet markets. Similarly, the polyprotein (orf1b, partial cds), spike (S) protein, envelop (E) protein, membrane (M) protein, non-structure protein 6 (NS6), nucleocapsid (N) protein and non-structure protein 7 (NS7) genes of HKU15 share 98.70% nucleotide identity with those of the δ-CoV (GenBank accession no. EF584908) detected from an Asian leopard cat [7]. More importantly, a recent retrospective study has confirmed the presence of PDCoV (CHN/AH/2004) in diarrheic pigs as early as 2004 in Anhui Province, China [19]. These facts suggest that these mammalian δ-CoVs directly or indirectly evolved from avian δ-CoVs. The transmission direction between the small wild carnivores and pigs is still unknown: (1) The wild small carnivore mammals could have been infected through catching and eating δ-CoV-positive birds [20] and could/can act as the intermediate hosts to transmit PDCoV to pigs; or (2) the wild small carnivore mammals could have been infected as they were fed PDCoV-positive pig viscera, which was used as carnivore food in wet markets (Haitao Xiu, personal communication). Natural PDCoV infection of these two mammalian species has not been reported since 2007, even with additional surveillance attempts [7], further supporting the second possibility, although it needs to be validated with more data in the future. Phylogenetic and recombination analyses suggest that PDCoV might have resulted from several recombination events involving δ-CoVs from several avian species, such as sparrow δ-CoVs HKU17 and ISU73347, and quail δ-CoV HKU30 (Figure 1) [8][21]. Recent studies indicate that aquatic birds may serve as natural reservoirs for δ-CoVs, while terrestrial birds and mammalian species may represent spill over hosts [22][23]. Sparrows, one of the most common birds at pig farms during the wintertime, late fall, and early spring, could promote the transmission of δ-CoVs to pigs. For example, bird droppings in the farms or in contaminated feed grains may be consumed by pigs [24].
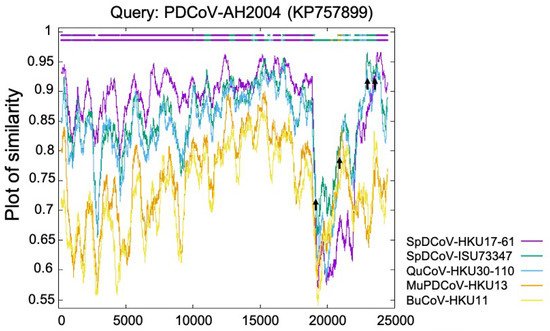
Figure 1. Identification of PDCoV-AH2004 strain as potential recombinant strain using Recombination Identification Program (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html, accessed on: 28 November 2021). At each position of the window, the query sequence PDCoV-AH2004 was compared with background sequences for 5 avian δ-CoVs (sparrow δ-CoVs HKU17 and ISU73347, quail δ-CoV HKU30, munia δ-CoV HKU13, and bulbul δ-CoV HKU11). The x-axis represents the length of the PDCoV genome, and the y-axis represents the similarity value. When the query sequence is like the background sequence, the homologous regions are indicated as thick dashed lines (of the corresponding color) on the top of the plot. Arrows represent potential recombination breakpoints.
Because the S gene of CoVs is the main genetic determinant of CoV host, tissue, or cellular tropism [25][26][27], Niu et al. [28] and Alhamo et al. [29] tested whether the spike protein or receptor-binding domain (RBD) from the sparrow δ-CoVs could alter PDCoV host and tissue tropism. First, Niu et al. [28] generated an infectious cDNA clone of PDCoV OH-FD22 strain (icPDCoV) and constructed two chimeric icPDCoVs harbouring the spike protein of HKU17 (icPDCoV-SHKU17) or the RBD of ISU73347 (icPDCoV-RBDISU). Niu et al. [28] and Alhamo et al. [29] evaluated their replication in the porcine kidney cell line LLC-PK1 and chicken fibroblast cell line DF-1 and performed pathogenesis studies in 4-day-old gnotobiotic pigs, 8-day-old turkey poults, and 11-day-old embryonated chicken eggs (ECEs). Compared with icPDCoV, the two chimeric viruses replicated to lower titers in LLC-PK1 cells. They did not cause clinical signs in pigs and replicated weakly in the nasal cavity but not in the intestines of pigs, whereas icPDCoV replicated mainly in the pig intestines and weakly in the respiratory tract. The two chimeric viruses and icPDCoV did not replicate in DF-1 cells at a multiplicity of infection (MOI) of 0.01 and did not replicate in the turkey poults or ECEs. Thus, these data indicate that the spike protein of sparrow δ-CoV HKU17 and the RBD of sparrow δ-CoV ISU73347 reduced PDCoV replication in pigs, suggesting limited potential for direct cross-species spillover from current sparrow strains to pigs. Recent molecular clock analyses suggest that PDCoVs shared a common ancestor with sparrow CoVs around 1810 and PDCoVs emerged around the 1990s [21].
Collectively, these studies suggest that PDCoV could have originated from terrestrial birds (e.g., sparrows) or less likely from other unknown wild mammalian δ-CoVs (Figure 2). Clearly, a better understanding of PDCoV origin requires detailed epidemiological studies of δ-CoVs in different species of birds and mammals.
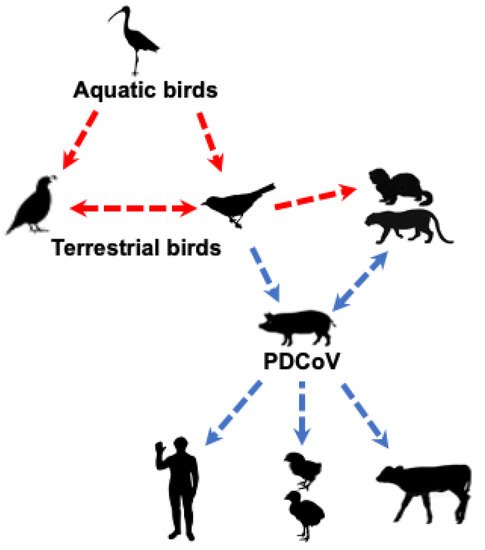
Figure 2. Likely origin and routes of cross-species transmission of PDCoV. The red dashed line indicates potential, but unknown, transmission of δ-CoVs from avian to mammalian species; the blue dashed line indicates potential transmission of PDCoV based on epidemiology or experimental studies.
3. Molecular Epidemiology and Genetic Diversity of PDCoVs
3.1. Prevalence of PDCoV in Different Countries
To date, PDCoV has been detected in Hong Kong, the United States, Canada, South Korea, mainland China, Thailand, Vietnam, Laos, Taiwan, Japan, Mexico and Haiti. The first documented PDCoV HKU15-44 and HKU15-155 strains were detected from the faecal samples collected from healthy pigs in 2009–2010 from a large-scale animal surveillance study in Hong Kong [7]. Subsequently, in early 2014, PDCoV associated diarrhoea outbreaks were first reported in three states of the US (Ohio, Iowa and Illinois). Later studies showed that PDCoV had spread to many Asian countries (China, Japan, Korea, Laos, Thailand, and Vietnam) and North American countries (the US, Canada, and Mexico) [10][30][31]. In the US, a retrospective surveillance study using reverse transcription-PCR (RT-qPCR) showed that PDCoV RNA was detected as early as August 2013 in pig faecal samples collected from Minnesota, Iowa and Illinois [32]. Furthermore, an indirect PDCoV S1 protein-based enzyme linked immunosorbent assay (ELISA) test suggested an even earlier presence of PDCoV IgG antibody in four archived serum samples collected in 2010 from US pigs [33]. In response to the large number of Swine Enteric Coronavirus Disease (SECD) cases since 2014, the United States Department of Agriculture (USDA) issued a federal order in June 2014, making SECD (caused by PEDV or PDCoV) reportable diseases, which was rescinded on 6 March 2018. According to the last SECD weekly situation report on 8 March 2018, 300 confirmed PDCoV positive premises and 88 presumptive positive premises cumulated between June 2014 and March 2018 and were distributed in 18 states within the US (https://www.aasv.org/pedv/SECD_Situation_Report_180308.pdf; accessed on: 15 October 2021). To date, PDCoV diarrhea occurs at a relatively low reported case rate according to the Swine Health Information Center (https://www.swinehealth.org/domestic-disease-surveillance-reports/; accessed on: 15 October 2021).
In mainland China, PDCoV was first reported in 2015 [19][34][35][36]; however, retrospective studies have shown that PDCoV could have emerged as a swine pathogen as early as 2004 [19]. A recent PDCoV surveillance study reported that 94 (13.07%) of 719 porcine diarrhoea samples collected from 18 provinces in China from March 2016 to June 2018 were PDCoV-positive by RT-qPCR [37]. The complete S genes of 11 PDCoV strains were determined. Seven were expectedly grouped into the Chinese lineage. However, CH-WH-2017 (GenBank accession no. MK040451) exhibited a closer relationship to the US/Japan/South Korea lineage; CH-HA3-2017, CH-HA1-2017 and CH-HA2-2017 strains (GenBank accession no. MK040455, MK040453 and MK040454) were clustered into a new branch and separate from the Chinese lineage. The latter showed the closest relationship to the Vietnam/Laos/Thailand lineage [37]. As of October 2021, PDCoV has been detected in 26 provinces of China, including all swine producing areas [35][37][38][39][40][41][42]. From mid-March 2014 to January 2016, PDCoV related diarrhoea outbreaks were successively identified in Canada, South Korea, Thailand, Vietnam, and Laos [30][43][44][45]. Retrospective studies revealed that PDCoV has been circulating in Taiwan [46], Japan [47], Mexico [48] and Haiti [18] since 2011, late 2013, 2015 and 2014, respectively. PDCoVs or PDCoV-like δ-CoVs have not been detected in the Europe, South American, African, and Australian continents, although retrospective studies have been conducted in Brazil [49] and Spain [50] to identify CoVs in wild birds and pigs, respectively.
3.2. Genetic Diversity of Global PDCoV
To date, 122 complete genome sequences of PDCoVs are available in GenBank (Table 1 and Figure 3). Phylogenetic analysis of these PDCoV genome sequences showed that all US, South Korean and Japanese strains clustered together; and three Chinese isolates (CHN-GD16-05/2016; CHzmd2019; CH-HLJ-20/2020) and one Haiti isolate (PDCoV/Haiti/Human/0256-1/2015) appeared to be more closely related to US/South Korean/Japanese strains. Therefore, these latter four strains were classified into the US linage (Figure 3). Similarly, He et al. found two other newly sequenced Chinese strains (AH2019/H and SD2019/426, GenBank accession no. are not available) were also clustered into the US lineage [10]. The close genetic relatedness between Chinese and US PDCoVs is not surprising due to the frequent trade of pork and pork products and pig feed supplements (https://www.fas.usda.gov/china-2020-export-highlights; accessed on: 10 November 2021). Additionally, it is not surprising to see that one of the three Haitian PDCoVs detected from children grouped within US lineage because pigs were re-introduced from the US to Haiti after the local pig population was mostly wiped out following the African Swine Fever epidemic in 1970s–1980s [51]. The early Chinese lineage contains the earliest strain that was detected from a sample collected in the Anhui Province in 2004 (CHN-AH-2004) and the strains isolated in Hong Kong in 2009 (HKU15-44) and 2010 (HKU15-155). The Chinese lineage is also inclusive of the two Haiti PDCoVs (PDCoV/Haiti/Human/0081-4/2014 and PDCoV/Haiti/Human/0329-4/2015) and the currently circulating strains in mainland China. The two Haitian strains are closely related to a Chinese strain (CHN/Tianjin/2016). According to the pork trade data from the Observatory of Economic Complexity (OEC), Chinese pork was exported to Haiti from 2006 to 2011 (https://oec.world/en/visualize/tree_map/hs92/import/hti/show/10203/2012/; accessed on: 10 November 2021). This may have contributed to PDCoV transmission from China to Haiti. The Southeast Asian (or Vietnam/Laos/Thailand) lineage contains PDCoV strains prevailing in these countries and a novel strain detected in China (CHN/GX/1468B/2017), indicating that these strains may derive from common evolutionary ancestors [52]. In addition, another Thailand-like PDCoV isolate (CHN-GX81-2018) was also discovered in Guangxi Province of China [53]. Collectively, phylogenetic analysis demonstrated that multiple PDCoV lineages, including US lineage, early Chinese lineage, Chinese lineage, and Vietnam/Laos/Thailand lineage, coexist in mainland China. He et al. [10] reported that more frequent intra- and inter-lineage recombination and higher virus genetic diversity were identified among and within the Vietnam/Laos/Thailand and Chinese lineages than within the US lineage. Frequent recombination events between different lineages of PDCoV strains in China may complicate PDCoV epidemiology and result in the emergence of PDCoV strains with changed pathogenicity and host tropism.
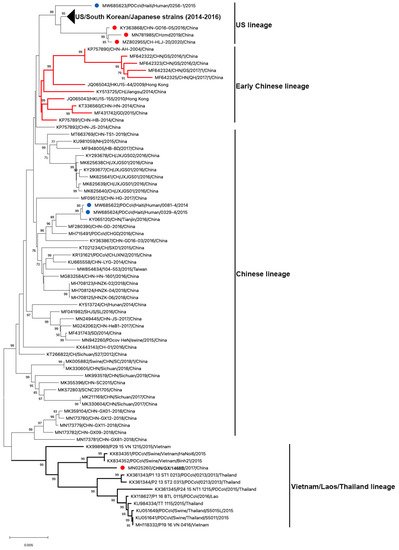
Figure 3. Phylogenetic analyses of PDCoV based on 122 complete genomes. Genome sequences were aligned with MAFFT v.7.490 [54]. The phylogenetic tree was constructed using the neighbour-joining method of MEGA 11, and bootstrap values (1000 replicates) above 70% are shown. The bar represents a corrected genetic distance. The red circles indicate PDCoV strains from China but that fall into non-Chinese lineages; the blue circles indicate PDCoV strains from humans in Haiti.
Table 1. Complete genome information on PDCoV strains.
| Lineage | GenBank Accession No. | Strain Name | Countries | Collection Date | Host | Reference |
|---|---|---|---|---|---|---|
| US | KJ462462.1 | OH1987 | USA | 31-Jan-2014 | Pig | [9] |
| KJ481931.1 | PDCoV/USA/Illinois121/2014 | USA | 04-Jan-2014 | Pig | [55] | |
| KJ567050.1 | 8734/USA-IA/2014 | USA | 20-Feb-2014 | Pig | [56] | |
| KJ569769.1 | IN2847 | USA | 13-Feb-2014 | Pig | [57] | |
| KJ584355.1 | IL2768 | USA | 12-Feb-2014 | Pig | [58] | |
| KJ584356.1 | SD3423 | USA | 20-Feb-2014 | Pig | [58] | |
| KJ584357.1 | KY4813 | USA | 07-Mar-2014 | Pig | [58] | |
| KJ584358.1 | PA3148 | USA | 18-Feb-2014 | Pig | [58] | |
| KJ584359.1 | NE3579 | USA | 21-Feb-2014 | Pig | [58] | |
| KJ620016.1 | MI6148 | USA | 18-Mar-2014 | Pig | [58] | |
| KM012168.1 | Michigan/8977/2014 | USA | 17-Mar-2014 | Pig | Unpublished | |
| KR150443.1 | USA/Arkansas61/2015 | USA | 24-Mar-2015 | Pig | Unpublished | |
| KR265847.1 | USA/Minnesota442/2014 | USA | 06-Mar-2014 | Pig | Unpublished | |
| KR265848.1 | USA/Minnesota214/2014 | USA | 14-Mar-2014 | Pig | Unpublished | |
| KR265849.1 | USA/Michigan447/2014 | USA | 02-Apr-2014 | Pig | Unpublished | |
| KR265850.1 | USA/Michigan448/2014 | USA | 02-Apr-2014 | Pig | Unpublished | |
| KR265851.1 | USA/Indiana453/2014 | USA | 13-May-2014 | Pig | Unpublished | |
| KR265852.1 | USA/Illinois449/2014 | USA | 21-Apr-2014 | Pig | Unpublished | |
| KR265853.1 | USA/Minnesota/2013 | USA | 14-Oct-2013 | Pig | Unpublished | |
| KR265854.1 | USA/Minnesota454/2014 | USA | 21-May-2014 | Pig | Unpublished | |
| KR265855.1 | USA/Minnesota455/2014 | USA | 21-May-2014 | Pig | Unpublished | |
| KR265856.1 | USA/Illinois272/2014 | USA | 23-Feb-2014 | Pig | Unpublished | |
| KR265857.1 | USA/Illinois273/2014 | USA | 23-Feb-2014 | Pig | Unpublished | |
| KR265858.1 | USA/NorthCarolina452/2014 | USA | 06-May-2014 | Pig | Unpublished | |
| KR265859.1 | USA/Minnesota159/2014 | USA | 11-Feb-2014 | Pig | Unpublished | |
| KR265860.1 | USA/Nebraska209/2014 | USA | 05-Feb-2014 | Pig | Unpublished | |
| KR265861.1 | USA/Nebraska210/2014 | USA | 05-Feb-2014 | Pig | Unpublished | |
| KR265862.1 | USA/Ohio444/2014 | USA | 26-Mar-2014 | Pig | Unpublished | |
| KR265863.1 | USA/Ohio445/2014 | USA | 27-Mar-2014 | Pig | Unpublished | |
| KR265864.1 | USA/Minnesota292/2014 | USA | 14-Mar-2014 | Pig | Unpublished | |
| KR265865.1 | USA/Iowa459/2014 | USA | 05-Jun-2014 | Pig | Unpublished | |
| KT381613.1 | OH11846 | USA | 07-May-2014 | Pig | [59] | |
| KX022602.1 | PDCoV/USA/Iowa136/2015 | USA | 15-Oct-2015 | Pig | Unpublished | |
| KX022603.1 | PDCoV/USA/Minnesota140/2015 | USA | 18-Dec-2015 | Pig | Unpublished | |
| KX022604.1 | PDCoV/USA/Nebraska137/2015 | USA | 27-Nov-2015 | Pig | Unpublished | |
| KX022605.1 | PDCoV/USA/Nebraska145/2015 | USA | 21-Dec-2015 | Pig | Unpublished | |
| MZ291567.1 | OH-FD22 P7 | USA | 2014 | Pig | [60] | |
| KY354363.1 | DH1 | South Korea | 01-Apr-2016 | Pig | [61] | |
| KY354364.1 | DH2 | South Korea | 01-Apr-2016 | Pig | [61] | |
| KY364365.1 | KNU16-07 | South Korea | Jul-2014 | Pig | [62] | |
| KY926512.1 | KNU16-11 | South Korea | Nov-2016 | Pig | [63] | |
| KM820765.1 | KNU14-04 | South Korea | Apr-2014 | Pig | [43] | |
| LC260038.1 | AKT/JPN/2014 | Japan | May-2014 | Pig | [47] | |
| LC260039.1 | GNM-1/JPN/2014 | Japan | May-2014 | Pig | [47] | |
| LC260040.1 | GNM-2/JPN/2014 | Japan | May-2014 | Pig | [47] | |
| LC260041.1 | IWT/JPN/2014 | Japan | May-2014 | Pig | [47] | |
| LC260042.1 | MYZ/JPN/2014 | Japan | May-2014 | Pig | [47] | |
| LC260043.1 | OKN/JPN/2014 | Japan | Aug-2014 | Pig | [47] | |
| LC260044.1 | YMG/JPN/2014 | Japan | Dec-2014 | Pig | [47] | |
| LC260045.1 | HKD/JPN/2016 | Japan | Sep-2016 | Pig | [64] | |
| KY363868.1 | CHN-GD16-05 | China | 05-Jan-2016 | Pig | [65] | |
| MN781985.1 | CHzmd2019 | China | Unknown | Pig | Unpublished | |
| MZ802955.1 | CH-HLJ-20 | China | Sep-2020 | Pig | Unpublished | |
| MW685623.1 | PDCoV/Haiti/Human/0256-1/2015 | Haiti | 16-Mar-2015 | Human | [18] | |
| Early Chinese |
KP757890.1 | CHN-AH-2004 | China | 24-May-2004 | Pig | [19] |
| KP757891.1 | CHN-HB-2014 | China | 26-Dec-2014 | Pig | [19] | |
| JQ065042.2 | HKU15-44 | China: Hong Kong | 2009 | Pig | [7] | |
| JQ065043.2 | HKU15-155 | China: Hong Kong | 2010 | Pig | [7] | |
| KT336560.1 | CHN-HN-2014 | China | 24-Nov-2014 | Pig | [66] | |
| KY513725.1 | CH/Jiangsu/2014 | China | 2014 | Pig | Unpublished | |
| MF431742.1 | GD | China | 2015 | Pig | Unpublished | |
| MF642322.1 | CHN/GS/2016/1 | China | Aug-2016 | Pig | [67] | |
| MF642323.1 | CHN/GS/2016/2 | China | Aug-2016 | Pig | [67] | |
| MF642324.1 | CHN/GS/2017/1 | China | Apr-2017 | Pig | [67] | |
| MF642325.1 | CHN/QH/2017/1 | China | Mar-2017 | Pig | [67] | |
| Chinese | KP757892.1 | CHN-JS-2014 | China | 20-Dec-2014 | Pig | [19] |
| KR131621.1 | PDCoV/CHJXNI2/2015 | China | Mar-2015 | Pig | [35] | |
| KT021234.1 | CH/SXD1/2015 | China | 20-Mar-2015 | Pig | [34] | |
| KT266822.1 | CH/Sichuan/S27/2012 | China | 2012 | Pig | [36] | |
| KU665558.1 | CHN-LYG-2014 | China | 26-Jun-2014 | Pig | Unpublished | |
| KU981059.1 | NH | China | 16-Feb-2015 | Pig | Unpublished | |
| KX443143.2 | CH-01 | China | 2016 | Pig | [68] | |
| KY065120.1 | CHN/Tianjin/2016 | China | 2016 | Pig | [69] | |
| KY293677.1 | CH/JXJGS01/2016 | China | 23-May-2016 | Pig | [70] | |
| KY293678.1 | CH/JXJGS02/2016 | China | 23-May-2016 | Pig | Unpublished | |
| KY363867.1 | CHN-GD16-03 | China | 18-Mar-2016 | Pig | [65] | |
| KY513724.1 | CH/Hunan/2014 | China | 2014 | Pig | Unpublished | |
| MF041982.1 | SHJS/SL/2016 | China | 23-Dec-2016 | Pig | [71] | |
| MF095123.1 | CHN-HG-2017 | China | 15-Feb-2017 | Pig | [72] | |
| MF280390.1 | CHN-GD-2016 | China | 2016 | Pig | [73] | |
| MF431743.1 | SD | China | 2014 | Pig | Unpublished | |
| MF948005.1 | HB-BD | China | 10-Aug-2017 | Pig | [74] | |
| MG242062.1 | CHN-HeB1-2017 | China | 2017 | Pig | Unpublished | |
| MG832584.1 | CHN-HN-1601 | China | Jul-2016 | Pig | [75] | |
| MH708123.1 | HNZK-02 | China | 20-Mar-2018 | Pig | [68] | |
| MH708124.1 | HNZK-04 | China | 20-Mar-2018 | Pig | [76] | |
| MH708125.1 | HNZK-06 | China | 20-Mar-2018 | Pig | [76] | |
| MH715491.1 | PDCoV/CHGD/2016 | China | 2016 | Pig | Unpublished | |
| MK005882.1 | Swine/CHN/SC/2018/1 | China | Mar-2018 | Pig | Unpublished | |
| MK211169.1 | CHN/Sichuan/2017 | China | 24-Dec-2017 | Pig | Unpublished | |
| MK330604.1 | CHN/Sichuan/2017 | China | Feb-2017 | Pig | Unpublished | |
| MK330605.1 | CHN/Sichuan/2018 | China | Jan-2018 | Pig | Unpublished | |
| MK355396.1 | CHN-SC2015 | China | 20-Feb-2016 | Pig | [77] | |
| MK359104.1 | CHN-GX01-2018 | China | 2018 | Pig | [53] | |
| MK572803.1 | SCNC201705 | China | Jun-2017 | Pig | Unpublished | |
| MK625638.1 | CH/JXJGS01/2016 | China | Oct-2018 | Pig | [70] | |
| MK625639.1 | CH/JXJGS01/2016 | China | Oct-2018 | Pig | [70] | |
| MK625640.1 | CH/JXJGS01/2016 | China | Oct-2018 | Pig | [70] | |
| MK625641.1 | CH/JXJGS01/2016 | China | Oct-2018 | Pig | [70] | |
| MK993519.1 | CHN/Sichuan/2019 | China | Jan-2019 | Pig | Unpublished | |
| MN173779.1 | CHN-GX11-2018 | China | 2018 | Pig | [53] | |
| MN173780.1 | CHN-GX12-2018 | China | 2018 | Pig | [53] | |
| MN173781.1 | CHN-GX81-2018 | China | 2018 | Pig | [53] | |
| MN173782.1 | CHN-GX09-2018 | China | 2018 | Pig | [53] | |
| MN249445.1 | CHN-JS-2017 | China | 11-Dec-2017 | Pig | [78] | |
| MN942260.1 | PDCoV HeN/swine/2015 | China | 2015 | Pig | Unpublished | |
| MW854634.1 | 104-553 | China: Taiwan | Jun-2015 | Pig | [46] | |
| MT663769.1 | CHN-TS1-2019 | China | 23-Jul-2019 | Pig | Unpublished | |
| MW685622.1 | PDCoV/Haiti/Human/0081-4/2014 | Haiti | 15-Dec-2014 | Human | [18] | |
| MW685624.1 | PDCoV/Haiti/Human/0329-4/2015 | Haiti | 13-Apr-2015 | Human | [18] | |
| Vietnam/ Laos/ Thailand |
KU051641.1 | PDCoV/Swine/Thailand/S5011/2015 | Thailand | 10-Jun-2015 | Pig | [79] |
| KU051649.1 | PDCoV/Swine/Thailand/S5015L/2015 | Thailand | 30-Jun-2015 | Pig | [79] | |
| KU984334.1 | TT_1115 | Thailand | Nov-2015 | Pig | [80] | |
| KX118627.1 | P1_16_BTL_0115/PDCoV/2016/Lao | Laos | 20-Jan-2016 | Pig | [44] | |
| KX361343.1 | P1_13_ST1_0213/PDCoV/0213/Thailand | Thailand | Feb-2013 | Pig | [81] | |
| KX361344.1 | P2_13_ST2_0313/PDCoV/0213/Thailand | Thailand | Mar-2013 | Pig | [81] | |
| KX361345.1 | P24_15_NT1_1215/PDCoV/2015/Thailand | Thailand | Dec-2015 | Pig | [81] | |
| KX834351.1 | PDCoV/Swine/Vietnam/HaNoi6/2015 | Vietnam | 10-Oct-2015 | Pig | [82] | |
| KX834352.1 | PDCoV/Swine/Vietnam/Binh21/2015 | Vietnam | 08-Dec-2015 | Pig | [82] | |
| KX998969.1 | P29_15_VN_1215 | Vietnam | Dec-2015 | Pig | Unpublished | |
| MH118332.1 | P19_16_VN_0416 | Vietnam | Unknown | Pig | [83] | |
| MN025260.1 | CH/GX/1468B/2017 | China | 18-Jan-2017 | Pig | [52] |
3.3. Co-Infection of PDCoV with Other Porcine Enteric Viruses
Clinical signs associated with PDCoV infection were found to be less severe than PEDV infection, and the PDCoV prevalence rate in diarrheic pigs is about 30% [7][9][30]. Nevertheless, co-infection with other enteric viral pathogens, such as PEDV, transmissible gastroenteritis virus (TGEV) and porcine rotavirus (PRV), are common and may lead to more severe clinical disease [30][37][41][84][85]. Recent surveys conducted in the United States, Mexico, South Korea and mainland China found that PEDV is the most frequent co-infecting pathogen with PDCoV, with a detection rate ranging from 4.73% to 54.10% (Table 2). Furthermore, except for the US, multiple-infection cases with both PDCoV and two other viruses (PEDV, TGEV, or PRV) were identified in other countries, with a detection rate ranging from 0.12% to 18.80% (Table 2). Additionally, a recent study found the co-infection of PDCoV with 4 to 7 other viral pathogens in diarrhoea samples collected in China from 2015 to 2018. These pathogens were porcine astrovirus (PAstV), porcine teschovirus (PTV), porcine sapelovirus (PSV), porcine enterovirus (PEV) 9/10, torque teno sus virus 2 (TTSuV-2), mammalian reovirus (MRV), porcine torovirus (PToV), porcine kobuvirus (PKV) or porcine bocavirus (PBoV) (Table 2).
Table 2. Co-infection of PDCoV with other porcine enteric viruses in pig farms or diarrheic samples from 2012 to 2018.
| Year | Country | Pathogens | Positive Farms/Samples (Positive Rate) |
References |
|---|---|---|---|---|
| 2014–2018 | United States | PDCoV + PEDV | 307 | [86] |
| 2014–2017 | Mexico | PDCoV + PEDV | 46 (54.1%) | [48] |
| PDCoV + TGEV | 9 (10.6%) | |||
| PDCoV + PEDV + TGEV | 16 (18.8%) | |||
| 2014–2016 | South Korea | PDCoV + PEDV | 43 (6.3%) | [63] |
| PDCoV + PRV | 19 (2.78%) | |||
| PDCoV + PEDV + PRV | 2 (0.29%) | |||
| 2016–2018 | China | PDCoV + PEDV | 34 (4.73%) | [37] |
| 2012–2018 | PDCoV + PEDV | 380 (46.74%) | [41] | |
| PDCoV + TGEV | 30 (3.69%) | |||
| PDCoV + PEDV + PRV | 3 (0.37%) | |||
| PDCoV + PEDV + TGEV | 1 (0.12%) | |||
| 2015–2018 | PDCoV + PAstV + PEV + TTSuV-2 + MRV + PKV | 1 (1.12%) | [87] | |
| PDCoV + PAstV + PEV + PKV + PBoV | 1 (1.12%) | |||
| PDCoV + PAstV + PTV + PSV + PEV + PKV + PBoV | 1 (1.12%) | |||
| PDCoV + PAstV + PTV + PSV + PKV + PBoV | 1 (1.12%) | |||
| PDCoV + PAstV + PTV + PSV + PEV + TTSuV-2 + PKV + PBoV | 1 (1.12%) | |||
| PDCoV + PAstV + PTV + PEV + TTSuV-2 + PToV + PBoV | 1 (1.12%) | |||
| PDCoV + PAstV + PEV + TTSuV-2 + PBoV | 1 (1.12%) |
This entry is adapted from the peer-reviewed paper 10.3390/pathogens11010079
References
- Geoghegan, J.L.; Duchene, S.; Holmes, E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017, 13, e1006215.
- Kenney, S.P.; Wang, Q.; Vlasova, A.; Jung, K.; Saif, L. Naturally Occurring Animal Coronaviruses as Models for Studying Highly Pathogenic Human Coronaviral Disease. Vet. Pathol. 2021, 58, 438–452.
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 163, 2601–2631.
- Dong, B.Q.; Liu, W.; Fan, X.H.; Vijaykrishna, D.; Tang, X.C.; Gao, F.; Li, L.F.; Li, G.J.; Zhang, J.X.; Yang, L.Q.; et al. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007, 81, 6920–6926.
- Chu, D.K.; Leung, C.Y.; Gilbert, M.; Joyner, P.H.; Ng, E.M.; Tse, T.M.; Guan, Y.; Peiris, J.S.; Poon, L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011, 85, 12815–12820.
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lai, K.K.; Huang, Y.; Lee, P.; Luk, G.S.; Dyrting, K.C.; Chan, K.H.; Yuen, K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009, 83, 908–917.
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008.
- Lau, S.K.P.; Wong, E.Y.M.; Tsang, C.C.; Ahmed, S.S.; Au-Yeung, R.K.H.; Yuen, K.Y.; Wernery, U.; Woo, P.C.Y. Discovery and Sequence Analysis of Four Deltacoronaviruses from Birds in the Middle East Reveal Interspecies Jumping with Recombination as a Potential Mechanism for Avian-to-Avian and Avian-to-Mammalian Transmission. J. Virol. 2018, 92, e00265-18.
- Wang, L.; Byrum, B.; Zhang, Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230.
- He, W.T.; Ji, X.; He, W.; Dellicour, S.; Wang, S.; Li, G.; Zhang, L.; Gilbert, M.; Zhu, H.; Xing, G.; et al. Genomic Epidemiology, Evolution, and Transmission Dynamics of Porcine Deltacoronavirus. Mol. Biol. Evol. 2020, 37, 2641–2654.
- Jung, K.; Hu, H.; Eyerly, B.; Lu, Z.; Chepngeno, J.; Saif, L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015, 21, 650–654.
- Chen, Q.; Gauger, P.; Stafne, M.; Thomas, J.; Arruda, P.; Burrough, E.; Madson, D.; Brodie, J.; Magstadt, D.; Derscheid, R.; et al. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 2015, 482, 51–59.
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49.
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016, 226, 50–59.
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States. Emerg. Infect. Dis. 2020, 26, 255–265.
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573.
- Jung, K.; Hu, H.; Saif, L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017, 162, 2357–2362.
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137.
- Dong, N.; Fang, L.; Zeng, S.; Sun, Q.; Chen, H.; Xiao, S. Porcine Deltacoronavirus in Mainland China. Emerg. Infect. Dis. 2015, 21, 2254–2255.
- Lo, C.L.C.; Fernandez, D.A.P.; de Luna, M.C.T.; de Guia, A.P.O.; Paller, V.G.V. Diet, parasites, and other pathogens of Sunda leopard cats (Prionailurus javanensis Desmarest 1816) in Aborlan, Palawan Island, Philippines. J. Parasit. Dis. 2021, 45, 627–633.
- Ye, X.; Chen, Y.; Zhu, X.; Guo, J.; Da, X.; Hou, Z.; Xu, S.; Zhou, J.; Fang, L.; Wang, D.; et al. Cross-Species Transmission of Deltacoronavirus and the Origin of Porcine Deltacoronavirus. Evol. Appl. 2020, 13, 2246–2253.
- Vlasova, A.N.; Kenney, S.P.; Jung, K.; Wang, Q.; Saif, L.J. Deltacoronavirus Evolution and Transmission: Current Scenario and Evolutionary Perspectives. Front. Vet. Sci. 2020, 7, 626785.
- Wille, M.; Holmes, E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020, 44, 631–644.
- Chen, Q.; Wang, L.; Yang, C.; Zheng, Y.; Gauger, P.C.; Anderson, T.; Harmon, K.M.; Zhang, J.; Yoon, K.J.; Main, R.G.; et al. The emergence of novel sparrow deltacoronaviruses in the United States more closely related to porcine deltacoronaviruses than sparrow deltacoronavirus HKU17. Emerg. Microbes Infect. 2018, 7, 105.
- Kuo, L.; Godeke, G.J.; Raamsman, M.J.; Masters, P.S.; Rottier, P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J. Virol. 2000, 74, 1393–1406.
- Haijema, B.J.; Volders, H.; Rottier, P.J. Switching species tropism: An effective way to manipulate the feline coronavirus genome. J. Virol. 2003, 77, 4528–4538.
- Sanchez, C.M.; Pascual-Iglesias, A.; Sola, I.; Zuniga, S.; Enjuanes, L. Minimum Determinants of Transmissible Gastroenteritis Virus Enteric Tropism Are Located in the N-Terminus of Spike Protein. Pathogens 2019, 9, 2.
- Niu, X.; Hou, Y.J.; Jung, K.; Kong, F.; Saif, L.J.; Wang, Q. Chimeric Porcine Deltacoronaviruses with Sparrow Coronavirus Spike Protein or the Receptor-Binding Domain Infect Pigs but Lose Virulence and Intestinal Tropism. Viruses 2021, 13, 122.
- Alhamo, M.A.; Boley, P.A.; Niu, X.; Yadav, K.K.; Lee, C.; Wang, Q.; Saif, L.; Kenney, S.P. Characterization of the Cross-Species Transmission Potential for Porcine Deltacoronaviruses Expressing Sparrow Coronavirus Spike Protein in Commercial Poultry. Viruses 2021. accepted.
- Marthaler, D.; Raymond, L.; Jiang, Y.; Collins, J.; Rossow, K.; Rovira, A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014, 20, 1347–1350.
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 2015, 6, e00064.
- Sinha, A.; Gauger, P.; Zhang, J.; Yoon, K.J.; Harmon, K. PCR-based retrospective evaluation of diagnostic samples for emergence of porcine deltacoronavirus in US swine. Vet. Microbiol. 2015, 179, 296–298.
- Thachil, A.; Gerber, P.F.; Xiao, C.T.; Huang, Y.W.; Opriessnig, T. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies. PLoS ONE 2015, 10, e0124363.
- Chen, F.; Zhu, Y.; Wu, M.; Ku, X.; Yao, L.; He, Q. Full-Length Genome Characterization of Chinese Porcine Deltacoronavirus Strain CH/SXD1/2015. Genome Announc. 2015, 3, e01284-15.
- Song, D.; Zhou, X.; Peng, Q.; Chen, Y.; Zhang, F.; Huang, T.; Zhang, T.; Li, A.; Huang, D.; Wu, Q.; et al. Newly Emerged Porcine Deltacoronavirus Associated with Diarrhoea in Swine in China: Identification, Prevalence and Full-Length Genome Sequence Analysis. Transbound Emerg. Dis. 2015, 62, 575–580.
- Wang, Y.W.; Yue, H.; Fang, W.; Huang, Y.W. Complete Genome Sequence of Porcine Deltacoronavirus Strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015, 3, e00945-15.
- Zhang, Y.; Cheng, Y.; Xing, G.; Yu, J.; Liao, A.; Du, L.; Lei, J.; Lian, X.; Zhou, J.; Gu, J. Detection and spike gene characterization in porcine deltacoronavirus in China during 2016–2018. Infect. Genet. Evol. 2019, 73, 151–158.
- Zhai, S.L.; Wei, W.K.; Li, X.P.; Wen, X.H.; Zhou, X.; Zhang, H.; Lv, D.H.; Li, F.; Wang, D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol. J. 2016, 13, 136.
- Li, D.; Feng, H.; Liu, Y.; Chen, Y.; Wei, Q.; Wang, J.; Liu, D.; Huang, H.; Su, Y.; Wang, D.; et al. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res. Vet. Sci. 2018, 120, 63–69.
- Li, B.; Zheng, L.; Li, H.; Ding, Q.; Wang, Y.; Wei, Z. Porcine deltacoronavirus causes diarrhea in various ages of field-infected pigs in China. Biosci. Rep. 2019, 39, BSR20190676.
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470.
- Tang, P.; Cui, E.; Song, Y.; Yan, R.; Wang, J. Porcine deltacoronavirus and its prevalence in China: A review of epidemiology, evolution, and vaccine development. Arch. Virol. 2021, 166, 2975–2988.
- Lee, S.; Lee, C. Complete Genome Characterization of Korean Porcine Deltacoronavirus Strain KOR/KNU14-04/2014. Genome Announc. 2014, 2, e01191-14.
- Lorsirigool, A.; Saeng-Chuto, K.; Temeeyasen, G.; Madapong, A.; Tripipat, T.; Wegner, M.; Tuntituvanont, A.; Intrakamhaeng, M.; Nilubol, D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 2016, 161, 2909–2911.
- Saeng-Chuto, K.; Lorsirigool, A.; Temeeyasen, G.; Vui, D.T.; Stott, C.J.; Madapong, A.; Tripipat, T.; Wegner, M.; Intrakamhaeng, M.; Chongcharoen, W.; et al. Different Lineage of Porcine Deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg. Dis. 2017, 64, 3–10.
- Hsu, T.H.; Liu, H.P.; Chin, C.Y.; Wang, C.; Zhu, W.Z.; Wu, B.L.; Chang, Y.C. Detection, sequence analysis, and antibody prevalence of porcine deltacoronavirus in Taiwan. Arch. Virol. 2018, 163, 3113–3117.
- Suzuki, T.; Shibahara, T.; Imai, N.; Yamamoto, T.; Ohashi, S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 2018, 61, 176–182.
- Perez-Rivera, C.; Ramirez-Mendoza, H.; Mendoza-Elvira, S.; Segura-Velazquez, R.; Sanchez-Betancourt, J.I. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg. Dis. 2019, 66, 1436–1441.
- Barbosa, C.M.; Durigon, E.L.; Thomazelli, L.M.; Ometto, T.; Marcatti, R.; Nardi, M.S.; de Aguiar, D.M.; Pinho, J.B.; Petry, M.V.; Neto, I.S.; et al. Divergent coronaviruses detected in wild birds in Brazil, including a central park in Sao Paulo. Braz. J. Microbiol. 2019, 50, 547–556.
- Puente, H.; Arguello, H.; Mencia-Ares, O.; Gomez-Garcia, M.; Rubio, P.; Carvajal, A. Detection and Genetic Diversity of Porcine Coronavirus Involved in Diarrhea Outbreaks in Spain. Front. Vet. Sci. 2021, 8, 651999.
- Alexander, F.C. Experiences with African swine fever in Haiti. Ann. N. Y. Acad. Sci. 1992, 653, 251–256.
- Wang, H.; Qin, Y.; Zhao, W.; Yuan, T.; Yang, C.; Mi, X.; Zhao, P.; Lu, Y.; Lu, B.; Chen, Z.; et al. Genetic Characteristics and Pathogenicity of a Novel Porcine Deltacoronavirus Southeast Asia-Like Strain Found in China. Front. Vet. Sci. 2021, 8, 701612.
- Huang, H.; Yin, Y.; Wang, W.; Cao, L.; Sun, W.; Shi, K.; Lu, H.; Jin, N. Emergence of Thailand-like strains of porcine deltacoronavirus in Guangxi Province, China. Vet. Med. Sci. 2020, 6, 854–859.
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492.
- Marthaler, D.; Jiang, Y.; Collins, J.; Rossow, K. Complete Genome Sequence of Strain SDCV/USA/Illinois121/2014, a Porcine Deltacoronavirus from the United States. Genome Announc. 2014, 2, e00218-14.
- Li, G.; Chen, Q.; Harmon, K.M.; Yoon, K.J.; Schwartz, K.J.; Hoogland, M.J.; Gauger, P.C.; Main, R.G.; Zhang, J. Full-Length Genome Sequence of Porcine Deltacoronavirus Strain USA/IA/2014/8734. Genome Announc. 2014, 2, e00278-14.
- Wang, L.; Zhang, Y.; Byrum, B. Complete Genome Sequence of Porcine Coronavirus HKU15 Strain IN2847 from the United States. Genome Announc. 2014, 2, e00291-14.
- Wang, L.; Byrum, B.; Zhang, Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014, 20, 1594–1595.
- Wang, L.; Hayes, J.; Sarver, C.; Byrum, B.; Zhang, Y. Porcine deltacoronavirus: Histological lesions and genetic characterization. Arch. Virol. 2016, 161, 171–175.
- Hu, H.; Jung, K.; Vlasova, A.N.; Chepngeno, J.; Lu, Z.; Wang, Q.; Saif, L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015, 53, 1537–1548.
- Chung, H.C.; Nguyen, V.G.; Oh, W.T.; My Le, H.T.; Moon, H.J.; Lee, J.H.; Kim, H.K.; Park, S.J.; Park, B.K. Complete Genome Sequences of Porcine Deltacoronavirus Strains DH1/2016 and DH2/2016 Isolated in South Korea. Genome Announc. 2017, 5, e01706-16.
- Jang, G.; Kim, S.H.; Lee, Y.J.; Kim, S.; Lee, D.S.; Lee, K.K.; Lee, C. Isolation and characterization of Korean porcine deltacoronavirus strain KNU16-07. J. Vet. Sci. 2018, 19, 577–581.
- Jang, G.; Lee, K.K.; Kim, S.H.; Lee, C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014–2016. Transbound Emerg. Dis. 2017, 64, 1364–1370.
- Suzuki, T.; Hayakawa, J.; Ohashi, S. Complete Genome Characterization of the Porcine Deltacoronavirus HKD/JPN/2016, Isolated in Japan, 2016. Genome Announc. 2017, 5, e00795-17.
- Mai, K.; Li, D.; Wu, J.; Wu, Z.; Cheng, J.; He, L.; Tang, X.; Zhou, Z.; Sun, Y.; Ma, J. Complete Genome Sequences of Two Porcine Deltacoronavirus Strains, CHN-GD16-03 and CHN-GD16-05, Isolated in Southern China, 2016. Genome Announc. 2018, 6, e01545-17.
- Dong, N.; Fang, L.; Yang, H.; Liu, H.; Du, T.; Fang, P.; Wang, D.; Chen, H.; Xiao, S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016, 196, 98–106.
- Wang, M.; Wang, Y.; Baloch, A.R.; Pan, Y.; Tian, L.; Xu, F.; Shivaramu, S.; Chen, S.; Zeng, Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound Emerg. Dis. 2018, 65, 363–369.
- Liang, Q.; Li, B.; Zhang, H.; Hu, H. Complete Genome Sequences of Two Porcine Deltacoronavirus Strains from Henan Province, China. Microbiol. Resour. Announc. 2019, 8, e01517-18.
- Zhao, F.; Sun, Y.; Qian, B.; Zhang, X.; Wu, Y. Complete Genome Characterization of Chinese Porcine Deltacoronavirus Strain CHN/Tianjin/2016. Genome Announc. 2017, 5, e00237-17.
- Zhang, M.; Ye, Y.; Gong, W.; Guo, N.; Zhang, F.; Li, A.; Zhou, X.; Huang, D.; Song, D.; Tang, Y. Complete Genome Sequence of Porcine Deltacoronavirus Strain CH/JXJGS01/2016, Isolated in Jiangxi Province, China, 2016. Genome Announc. 2017, 5, e00832-17.
- Yang, D.; Ju, H.; Wang, J.; Bai, Y.; Ge, F.; Liu, J.; Li, X.; Sun, Q.; Yang, X.C.; Zhu, J.; et al. Genome sequencing and analysis of a porcine delta coronavirus from eastern China. Eur. J. Exp. Biol. 2017, 7, 25.
- Zhang, M.J.; Liu, D.J.; Liu, X.L.; Ge, X.Y.; Jongkaewwattana, A.; He, Q.G.; Luo, R. Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Arch. Virol. 2019, 164, 413–425.
- Xu, Z.; Zhong, H.; Zhou, Q.; Du, Y.; Chen, L.; Zhang, Y.; Xue, C.; Cao, Y. A Highly Pathogenic Strain of Porcine Deltacoronavirus Caused Watery Diarrhea in Newborn Piglets. Virol. Sin. 2018, 33, 131–141.
- Liu, B.J.; Zuo, Y.Z.; Gu, W.Y.; Luo, S.X.; Shi, Q.K.; Hou, L.S.; Zhong, F.; Fan, J.H. Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound Emerg. Dis. 2018, 65, 874–882.
- Zhou, X.; Zhou, L.; Zhang, P.; Ge, X.; Guo, X.; Han, J.; Zhang, Y.; Yang, H. A strain of porcine deltacoronavirus: Genomic characterization, pathogenicity and its full-length cDNA infectious clone. Transbound Emerg. Dis. 2021, 68, 2130–2146.
- Zhang, H.; Liang, Q.; Li, B.; Cui, X.; Wei, X.; Ding, Q.; Wang, Y.; Hu, H. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev. Vet. Med. 2019, 166, 8–15.
- Zhao, Y.; Qu, H.; Hu, J.; Fu, J.; Chen, R.; Li, C.; Cao, S.; Wen, Y.; Wu, R.; Zhao, Q.; et al. Characterization and Pathogenicity of the Porcine Deltacoronavirus Isolated in Southwest China. Viruses 2019, 11, 1074.
- Qian, S.; Jia, X.; Gao, Z.; Zhang, W.; Xu, Q.; Li, Z. Isolation and Identification of Porcine Deltacoronavirus and Alteration of Immunoglobulin Transport Receptors in the Intestinal Mucosa of PDCoV-Infected Piglets. Viruses 2020, 12, 79.
- Janetanakit, T.; Lumyai, M.; Bunpapong, N.; Boonyapisitsopa, S.; Chaiyawong, S.; Nonthabenjawan, N.; Kesdaengsakonwut, S.; Amonsin, A. Porcine Deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016, 22, 757–759.
- Madapong, A.; Saeng-Chuto, K.; Lorsirigool, A.; Temeeyasen, G.; Srijangwad, A.; Tripipat, T.; Wegner, M.; Nilubol, D. Complete Genome Sequence of Porcine Deltacoronavirus Isolated in Thailand in 2015. Genome Announc. 2016, 4, e00408-16.
- Saeng-Chuto, K.; Stott, C.J.; Wegner, M.; Senasuthum, R.; Tantituvanont, A.; Nilubol, D. Retrospective investigation and evolutionary analysis of a novel porcine deltacoronavirus strain detected in Thailand from 2008 to 2015. Arch. Virol. 2017, 162, 2103–2108.
- Le, V.P.; Song, S.; An, B.H.; Park, G.N.; Pham, N.T.; Le, D.Q.; Nguyen, V.T.; Vu, T.T.H.; Kim, K.S.; Choe, S.; et al. A novel strain of porcine deltacoronavirus in Vietnam. Arch. Virol. 2018, 163, 203–207.
- Saeng-Chuto, K.; Jermsutjarit, P.; Stott, C.J.; Vui, D.T.; Tantituvanont, A.; Nilubol, D. Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound Emerg. Dis. 2020, 67, 183–198.
- Saeng-Chuto, K.; Madapong, A.; Kaeoket, K.; Pineyro, P.E.; Tantituvanont, A.; Nilubol, D. Coinfection of porcine deltacoronavirus and porcine epidemic diarrhea virus increases disease severity, cell trophism and earlier upregulation of IFN-alpha and IL12. Sci. Rep. 2021, 11, 3040.
- Zhang, H.; Han, F.; Shu, X.; Li, Q.; Ding, Q.; Hao, C.; Yan, X.; Xu, M.; Hu, H. Co-infection of porcine epidemic diarrhoea virus and porcine deltacoronavirus enhances the disease severity in piglets. Transbound Emerg. Dis. 2021.
- Animal and Plant Health Inspection Service. Available online: https://www.aasv.org/pedv/SECD_Situation_Report_180308.pdf (accessed on 15 October 2021).
- Su, M.; Qi, S.; Yang, D.; Guo, D.; Yin, B.; Sun, D. Coinfection and Genetic Characterization of Porcine Astrovirus in Diarrheic Piglets in China from 2015 to 2018. Front. Vet. Sci. 2020, 7, 462.
This entry is offline, you can click here to edit this entry!
