Flaviviruses are arthropod-borne RNA viruses that have been used extensively to study host antiviral responses. Often selected just to represent standard single-stranded positive-sense RNA viruses in early studies, the Flavivirus genus over time has taught us how truly unique it is in its remarkable ability to target not just the RNA sensory pathways but also the cytosolic DNA sensing system for its successful replication inside the host cell. This review summarizes the main developments on the unexpected antagonistic strategies utilized by different flaviviruses, with RNA genomes, against the host cyclic GAMP synthase (cGAS)/stimulator of interferon genes (STING) cytosolic DNA sensing pathway in mammalian systems. On the basis of the recent advancements on this topic, we hypothesize that the mechanisms of viral sensing and innate immunity are much more fluid than what we had anticipated, and both viral and host factors will continue to be found as important factors contributing to the host innate immune system in the future.
- flavivirus
- innate immunity
- pathogen sensing
- viral antagonism
- DNA sensing
1. Introduction
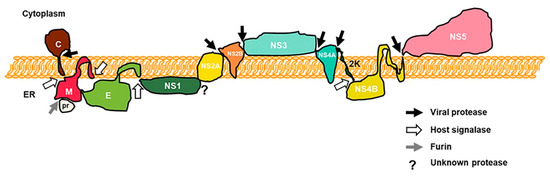
2. Innate Immune System
2.1. Overview
2.2. Viral Nucleic Acid Sensing
2.3. Innate Immune DNA Sensors
2.4. cGAS/STING Pathway
| Genus | Species | Protein | Mechanism of cGAS/STING Antagonism | References |
|---|---|---|---|---|
| Flavivirus | DENV | NS2B | Targets cGAS for degradation through an autophagy–lysosome-dependent mechanism. | [82] |
| NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [10,11,83,84,85,86] | ||
| ZIKV | NS1 | Indirectly degrades cGAS via the enhanced stabilization of caspase-1. | [87] | |
| NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | ||
| WNV | NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | |
| YFV | NS4B | Unknown | [19] | |
| JEV | NS2B3 | Proteolytically degrades human STING, but not mouse STING. | [85] | |
| DTMUV * | NS2A | Competes with duck TBK1 to bind with duck STING, thus disrupting its dimerization and inhibiting downstream IFN production. | [88] | |
| NS2B3 | Proteolytically degrades duck STING through an NS2B dependent manner with the binding of NS2B with duck STING being required for cleaving | [89] |
3. Perspectives
This entry is adapted from the peer-reviewed paper 10.3390/v12090979
