Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Cardiac arrhythmia, or irregular heart rhythm, is associated with morbidity and mortality and is described as one of the most important future public health challenges. In the last few decades, the zebrafish has emerged as an attractive model to reproduce in vivo human cardiac pathologies, including arrhythmias. As genetic tools in zebrafish continue to bloom, this model will be crucial for functional genomics studies and to develop personalized anti-arrhythmic therapies.
- cardiac arrhythmia
- heart development
- zebrafish
- atrial fibrillation
- cardiomyopathy
- imaging
- cardiac rhythm phenotyping
1. Introduction
The healthy human heart beats with a coordinated rhythm. Abnormal heart rhythm, or arrhythmia, refers to conditions in which heart rate or rhythmicity are altered or chaotic. The main inherited cardiac arrhythmias are long QT syndrome (LQTS), short QT syndrome (SQTS), catecholaminergic polymorphic ventricular tachycardia (CPVT), and Brugada syndrome (BrS). These diseases often result from mutations in genes encoding ion channels, leading to altered ionic currents that influence the cardiac action potential [1][2]. The most common cardiac arrhythmia is atrial fibrillation (AF), which is frequently associated with aging, inflammation, or following surgery [3][4][5]. A portion of AF cases arise in the absence of predisposing factors, often with a younger age of onset and with significant heritability [6]. Arrhythmias also occur in conjunction with inherited cardiomyopathies, including hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic cardiomyopathy (ACM), and left ventricular non-compaction cardiomyopathy (LVNC). These cardiomyopathies are frequently associated with mutations in genes encoding sarcomeric, desmosomal, or cytoskeletal proteins.
While often considered purely electrical diseases, primary cardiac arrhythmias have variable aetiologies. Genome-wide association studies (GWAS) and whole exome/genome sequencing techniques have implicated diverse pathways in the pathogenesis of cardiac arrhythmia, including developmental [7] and structural genes [8][9][10]. Many of the loci identified in GWAS are found in non-coding regions, suggesting that these variants alter gene expression which confers disease susceptibility [11][12].
The zebrafish has emerged as an exceptionally powerful model to study cardiac development and disease. From a practical perspective, zebrafish are optically transparent and develop externally, enabling observation during development. They have high fecundity such that large numbers of embryos are easily acquired. Manipulating the zebrafish genome is relatively straightforward, and numerous reporter lines allow for visualization of cellular and organ-level morphology and physiology. Notably, the small size of zebrafish enables them to survive without a functional cardiovascular system early in development, as their oxygen and nutritional needs can be met by diffusion into the embryo. This advantage permits analyses of mutants with little to no cardiac function [13], whereas orthologous mutants in other vertebrate models would not survive long enough to be observed.
2. Heart Development in Zebrafish
Heart development in zebrafish involves several distinct steps. This section will briefly outline the major processes that occur during each stage of heart development (Figure 1).
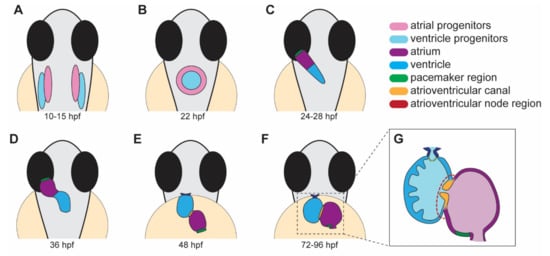
Figure 1. Stages of heart development in zebrafish from 10 to 96 h post-fertilization (hpf). (A) Atrial and ventricular cardiac progenitors are located in the anterior lateral plate mesoderm by ~15 hpf. (B) The cardiac disc is visible by 22 hpf as cardiac progenitors surround endocardial cells at the midline. (C) At 24 hpf, the linear heart tube forms and jogs to the left in preparation for heart looping. (D) At 36 hpf, the heart tube undergoes rightward looping. The AV canal (orange) begins to develop between the cardiac chambers. (E) At 48 hpf, the cardiac chambers begin to balloon and expand outwards. The bulbus arteriosus (dark blue) and AV canal (orange) continue to develop and mature. (F) From 72 to 96 hpf, the cardiac chambers expand and align beside each other. (G) A cross-section of a 96 hpf heart showing the trabeculae, the finger-like muscular projections on the inner wall of the ventricle, and endocardial leaflets (orange) of the AV canal. (A–D) dorsal views; (E–G) ventral views.
3. The Zebrafish Conduction System
The CCS is composed of pacemaker cells in the SAN, the atrioventricular node (AVN), and the fast ventricular conduction system. Although the zebrafish heart is formed by only a single atrium and single ventricle, similarities between its CCS with the human CCS supports the use of zebrafish as a model to study CCS development and function/dysfunction. Pacemaker cells have been identified in the sinus venosus and atrium junction [14], and slow-conducting AVC cardiomyocytes in the AVN region have been mapped in zebrafish [15]. While the mammalian His-Purkinje system is absent in zebrafish, the ventricular trabecular myocardium has been postulated to serve as a functional equivalent [16].
The molecular profiles of conducting tissues are highly conserved in zebrafish. Tomo-seq, a technique to spatially resolve genome-wide transcriptomics data, was used to profile the 2 days post-fertilization (dpf) zebrafish heart. These data identified a sub-compartment that highly expresses pacemaker development genes, including isl1 and shox2 [17]. Recent transcriptome profiling of the sinoatrial ring [18] and AVC cells [19] from the developing zebrafish heart confirms conserved gene expression signatures found in the mammalian SAN and AVN, respectively. Many of the core mammalian SAN/AVN genes are expressed in the developing zebrafish, including tbx18, hcn4, bmp4, cacna1ab in the sinoatrial ring [18], and an abundance of genes encoding connexins, T-type Ca2+ channel (cacna1g), and the pacemaker hyperpolarization channel (hcn4) in the AVN [19].
Cardiac physiology in zebrafish aligns closely with mammalian models. The average resting heart rate of humans is 60–90 beats per minute (bpm), while the average heart rate of zebrafish is 120–180 bpm [20]. This characteristic is a considerable advantage over the widely used rodent models like mouse, which have average heart rates of 300–600 bpm. Adult zebrafish basal electrocardiogram (ECG) characteristics are similar to humans, with a distinct P wave, QRS complex, and T-wave [21][22]. As in humans and large animal models, zebrafish have chamber-specific differences in action potential (AP) shape and duration (for examples, please see [21][23][24][25]). In atrial and ventricular cardiomyocytes of zebrafish, the resting membrane potential and AP amplitude are comparable with those observed in humans. Like humans, a clear plateau phase is established in ventricular APs, although a fast phase-1 repolarization is not present. Due to the elevated heart rate in zebrafish, the AP duration in the atrial and ventricular cardiomyocytes is shortened when compared to humans [25].
4. Zebrafish Models of Cardiac Arrhythmia
Cardiac arrhythmias have multifactorial aetiologies. Several zebrafish models with cardiac rhythm phenotypes have been reported (Table 1). Notably, these models provide unique insight toward disease initiation and mechanisms of pathogenesis.
Table 1. Zebrafish models of cardiac arrhythmia.
| Model/ Gene |
Allele | Cardiac Defect | Clinical Arrhythmia |
Human Ortholog | Ref. |
|---|---|---|---|---|---|
| atp1a1a.1 | hiphop (tx218) | 3:1 ratio of atrial contraction to ventricular contraction, bradycardia, and AV-block. |
LQTS | ATP1A1 | [26][27] |
| cacna1c | island beat (m379, m458, m231) |
Silent ventricle, uncoordinated contraction of the atrium. |
AF | CACNA1C | [13][28] |
| cmlc1 myl4 |
s977 bw24 |
Bradycardia, slow conduction in enlarged atrium, sarcomere disorganization. |
AF | MYL4 | [29][30] |
| cx43 (gja1b) | Morpholino | Bradycardia, AV-block, and fibrillation. |
AF | GJA1 | [31] |
| foxn4 | slipjig s644) | Peristaltic contraction with no AV delay. |
FOXN4 | [32][33] | |
| gja3/cx46 | dococ (s215, s226) | Uncoordinated conduction and contraction within the ventricle. |
CX46 | [34] | |
| hcn4 | Morpholino | Bradycardia and prolonged cardiac pauses. |
SSS | HCN4 | [35] |
| isl1 (K88X mutant) | sa0029 | 2 dpf: bradycardia due to impaired SA node function. 3–4 dpf: sinus block. |
SSS | ISL1 | [14][36] |
| kcnh6a (zerg) | breakdance (tb218) | 2:1 ratio of atrial to ventricular contraction, bradycardia, reduced cardiac output, and AV-block due to impairment of IKr channel. |
LQTS | KCNH6 (hERG) | [26][37] |
| kcnh6a (zerg) | reggae | Intermittent atrial fibrillation and acceleration of cardiomyocyte repolarization. |
SQTS | KCNH6 (hERG) | [38] |
| kcnma1b | Morpholino | Decreased contraction of heart chambers, sinus bradycardia. |
AF | KCNMA1 | [39] |
| mcu | la2446 | Cardiomyopathy. Thin, dilated atrium, small ventricle with restricted blood flow, swollen mitochondria. Heart rate variability. | SSS | MCU | [40] |
| nkx2.5 | vu176, vu413 | Reduced heart rate variation, increased heart rate. | CHD | NKX2-5 | [41] |
| pitx2c | ups6 | Embryonic: arrhythmia, sarcomere disorganization, increased ROS. Adult: extended P-wave and PR-interval, fibrosis, sarcomere disorganization. |
AF | PITX2 | [42] |
| pln | hu10742 | Adult: structural remodeling, immune cell infiltration, contractile defects, AP alternans, altered Ca2+ handling | ACM | PLN | [43][44] |
| scn5a | human variant |
Bradycardia, sinus pauses, AV-block. | LQTS | SCN5A | [45] |
| slc8a1a (ncx1) |
tremblor (tc318d, te381b, m116, m139, m158, m276, m736) | Fibrillation from onset of contraction (more prominent in the atrium than the ventricle). Absent circulation. | SLC8A1 (NCX1) |
[13][26][46] | |
| tbx5a | heartstrings (m21) | Slight bradycardia evident during initial heart tube stage. Heart fails to loop, contractility declines, and pericardial edema develops. |
Holt–Oram syndrome | TBX5 | [47] |
| tcf2 | hobgoblin (s634) | AV block at 48 hpf, silent ventricle at 96 hpf. |
TCF2 | [32] | |
| tmem161b | grime (uq4ks) | Bradycardia, skipped ventricular beats, increased heart rate variability | LQTS | TMEM161B | [48] |
| ttn.2 | sfc9 | Atrial fibrosis, compromised sarcomere assembly in atrium and ventricle, lengthened PR interval. |
AF | TTN | [49] |
| mobitz (s466) | AV block, sinus pause at 120 hpf. | [32] | |||
| elektra (s587) | AV block. | [32] | |||
| daredevil (s275, s563) | AV block, silent ventricle at 120 hpf. | [32] | |||
| bullseye (s885) | No heartbeat at 24 hpf, AV block at 36–48 hpf. |
[32] | |||
| kingpin (s886) | Atrial and ventricular fibrillation | [32] |
5. Techniques for Assessing Cardiac Rhythm and Function in Embryonic and Adult Zebrafish
5.1. Tools to Study Cardiac Rhythm at Embryonic Stages
Due to its amenability to live imaging and genetic manipulation, the zebrafish model provides a great opportunity for understanding the genetic and molecular mechanisms underlying cardiac arrhythmia.
The zebrafish embryo is easy to image due to its transparency, and light microscopy is suitable to detect early arrhythmia. High-speed acquisition of the beating heart is key to the identification of arrhythmia. One of these techniques, spinning disk microscopy, provides many attractive advantages for imaging heart contractions in vivo during development due to its speed with higher frame rates. The high speed allows the imaging of multiple samples in a short amount of time, making it an excellent instrument for high-throughput chemical screening assay in zebrafish embryos [50]. After acquiring heartbeat movies, a kymograph, a plot representing spatial position over time, can be generated to quantify heart rate, heart rate variability, and cardiac output in larvae [14][42][48][51].
Light-sheet microscopy is another fluorescence microscopy technique suitable to detect arrhythmia. It uses a plane of light to optically section and views tissues at a cellular resolution. Light-sheet microscopy presents the advantage of deep imaging with a thin plane of light, limiting phototoxicity and photobleaching [52]. By combining this approach with optogenetics, a technique in which channels can be controlled with light, Arrenberg et al. created an optically controlled pacemaker by expressing halorhodopsin and channelrhodopsin in zebrafish cardiomyocytes [15]. Using these tools, the cardiac pacemaker was mapped using a patterned illumination to localize the areas sensitive to hyperpolarization at the inflow tract and AV canal during early development.
5.2. Applications for Adult Cardiac Rhythm Phenotyping
Cardiac rhythm can be monitored in adult zebrafish even though it presents more challenges due to the opacity of the animal. Echocardiography is an ultrasound imaging method that uses a high-frequency transducer directly applied to the body of the anesthetized zebrafish. This technique is non-invasive and enables quantification of cardiac output parameters including chamber area, fractional area change, and fractional shortening using brightness mode (B-mode) [42]. Pulsed-wave Doppler imaging provides information on blood flow and valve function [53][54]. ECG is the most common method to evaluate cardiac electrophysiology clinically and may be applied in zebrafish. Electrodes are placed on top of the cardiac region to record the T wave, P wave, and QRS complex. The measurement of the P wave duration and PR intervals, representing atrial depolarization, can indicate atrial conduction defects, as has been reported for several models [21][42][49].
This entry is adapted from the peer-reviewed paper 10.3390/jcdd9010013
References
- Campuzano, O.; Beltrán-Álvarez, P.; Iglesias, A.; Scornik, F.; Pérez, G.; Brugada, R. Genetics and cardiac channelopathies. Genet. Med. 2010, 12, 260–267.
- Campuzano, O.; Brugada, R.; Iglesias, A. Genetics of Brugada syndrome. Curr. Opin. Cardiol. 2010, 25, 210–215.
- Heijman, J.; Muna, A.P.; Veleva, T.; Molina, C.E.; Sutanto, H.; Tekook, M.; Wang, Q.; Abu-Taha, I.H.; Gorka, M.; Künzel, S.; et al. Atrial Myocyte NLRP3/CaMKII Nexus Forms a Substrate for Postoperative Atrial Fibrillation. Circ. Res. 2020, 127, 1036–1055.
- Nattel, S.; Heijman, J.; Zhou, L.; Dobrev, D. Molecular Basis of Atrial Fibrillation Pathophysiology and Therapy: A Translational Perspective. Circ. Res. 2020, 127, 51–72.
- Dobrev, D.; Aguilar, M.; Heijman, J.; Guichard, J.B.; Nattel, S. Postoperative atrial fibrillation: Mechanisms, manifestations and management. Nat. Rev. Cardiol. 2019, 16, 417–436.
- Ragab, A.A.Y.; Sitorus, G.D.S.; Brundel, B.; de Groot, N.M.S. The Genetic Puzzle of Familial Atrial Fibrillation. Front. Cardiovasc. Med. 2020, 7, 14.
- Nielsen, J.B.; Thorolfsdottir, R.B.; Fritsche, L.G.; Zhou, W.; Skov, M.W.; Graham, S.E.; Herron, T.J.; McCarthy, S.; Schmidt, E.M.; Sveinbjornsson, G.; et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018, 50, 1234–1239.
- Andreasen, L.; Bertelsen, L.; Ghouse, J.; Lundegaard, P.R.; Ahlberg, G.; Refsgaard, L.; Rasmussen, T.B.; Eiskjær, H.; Haunsø, S.; Vejlstrup, N.; et al. Early-onset atrial fibrillation patients show reduced left ventricular ejection fraction and increased atrial fibrosis. Sci. Rep. 2020, 10, 10039.
- Goette, A.; Kalman, J.M.; Aguinaga, L.; Akar, J.; Cabrera, J.A.; Chen, S.A.; Chugh, S.S.; Corradi, D.; D’Avila, A.; Dobrev, D.; et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: Definition, characterization, and clinical implication. Heart Rhythm 2017, 14, e3–e40.
- Gudbjartsson, D.F.; Holm, H.; Sulem, P.; Masson, G.; Oddsson, A.; Magnusson, O.T.; Saemundsdottir, J.; Helgadottir, H.T.; Helgason, H.; Johannsdottir, H.; et al. A frameshift deletion in the sarcomere gene MYL4 causes early-onset familial atrial fibrillation. Eur. Heart J. 2017, 38, 27–34.
- Van Ouwerkerk, A.F.; Hall, A.W.; Kadow, Z.A.; Lazarevic, S.; Reyat, J.S.; Tucker, N.R.; Nadadur, R.D.; Bosada, F.M.; Bianchi, V.; Ellinor, P.T.; et al. Epigenetic and Transcriptional Networks Underlying Atrial Fibrillation. Circ. Res. 2020, 127, 34–50.
- Roselli, C.; Rienstra, M.; Ellinor, P.T. Genetics of Atrial Fibrillation in 2020: GWAS, Genome Sequencing, Polygenic Risk, and Beyond. Circ. Res. 2020, 127, 21–33.
- Stainier, D.Y.; Fouquet, B.; Chen, J.N.; Warren, K.S.; Weinstein, B.M.; Meiler, S.E.; Mohideen, M.A.; Neuhauss, S.C.; Solnica-Krezel, L.; Schier, A.F.; et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 1996, 123, 285–292.
- Tessadori, F.; van Weerd, J.H.; Burkhard, S.B.; Verkerk, A.O.; de Pater, E.; Boukens, B.J.; Vink, A.; Christoffels, V.M.; Bakkers, J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE 2012, 7, e47644.
- Arrenberg, A.B.; Stainier, D.Y.; Baier, H.; Huisken, J. Optogenetic control of cardiac function. Science 2010, 330, 971–974.
- Sedmera, D.; Reckova, M.; de Almeida, A.; Sedmerova, M.; Biermann, M.; Volejnik, J.; Sarre, A.; Raddatz, E.; McCarthy, R.A.; Gourdie, R.G.; et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am. J. Physiol. Heart Circ. Physiol. 2003, 284, H1152–H1160.
- Burkhard, S.B.; Bakkers, J. Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/beta-catenin signaling in autonomic control of heart rate. eLife 2018, 7, e31515.
- Minhas, R.; Loeffler-Wirth, H.; Siddiqui, Y.H.; Obrębski, T.; Vashisht, S.; Nahia, K.A.; Paterek, A.; Brzozowska, A.; Bugajski, L.; Piwocka, K.; et al. Transcriptome profile of the sinoatrial ring reveals conserved and novel genetic programs of the zebrafish pacemaker. BMC Genom. 2021, 22, 715.
- Abu Nahia, K.; Migdał, M.; Quinn, T.A.; Poon, K.-L.; Łapiński, M.; Sulej, A.; Liu, J.; Mondal, S.S.; Pawlak, M.; Bugajski, Ł.; et al. Genomic and physiological analyses of the zebrafish atrioventricular canal reveal molecular building blocks of the secondary pacemaker region. Cell. Mol. Life Sci. 2021, 78, 6669–6687.
- Sampurna, B.P.; Audira, G.; Juniardi, S.; Lai, Y.-H.; Hsiao, C.-D. A Simple ImageJ-Based Method to Measure Cardiac Rhythm in Zebrafish Embryos. Inventions 2018, 3, 21.
- Zhao, Y.; Yun, M.; Nguyen, S.A.; Tran, M.; Nguyen, T.P. In Vivo Surface Electrocardiography for Adult Zebrafish. J. Vis. Exp. 2019, 150, e60011.
- Milan, D.J.; Jones, I.L.; Ellinor, P.T.; MacRae, C.A. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H269–H273.
- Echeazarra, L.; Hortigón-Vinagre, M.P.; Casis, O.; Gallego, M. Adult and Developing Zebrafish as Suitable Models for Cardiac Electrophysiology and Pathology in Research and Industry. Front. Physiol. 2021, 11, 1692.
- Zhao, Y.; Chen, C.; Yun, M.; Issa, T.; Lin, A.; Nguyen, T.P. Constructing Adult Zebrafish Einthoven’s Triangle to Define Electrical Heart Axes. Front. Physiol. 2021, 12, 708938.
- Nemtsas, P.; Wettwer, E.; Christ, T.; Weidinger, G.; Ravens, U. Adult zebrafish heart as a model for human heart? An electrophysiological study. J. Mol. Cell. Cardiol. 2010, 48, 161–171.
- Chen, J.N.; Haffter, P.; Odenthal, J.; Vogelsang, E.; Brand, M.; van Eeden, F.J.; Furutani-Seiki, M.; Granato, M.; Hammerschmidt, M.; Heisenberg, C.P.; et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development 1996, 123, 293–302.
- Pott, A.; Bock, S.; Berger, I.M.; Frese, K.; Dahme, T.; Keßler, M.; Rinné, S.; Decher, N.; Just, S.; Rottbauer, W. Mutation of the Na(+)/K(+)-ATPase Atp1a1a.1 causes QT interval prolongation and bradycardia in zebrafish. J. Mol. Cell. Cardiol. 2018, 120, 42–52.
- Rottbauer, W.; Baker, K.; Wo, Z.G.; Mohideen, M.A.; Cantiello, H.F.; Fishman, M.C. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev. Cell 2001, 1, 265–275.
- Orr, N.; Arnaout, R.; Gula, L.J.; Spears, D.A.; Leong-Sit, P.; Li, Q.; Tarhuni, W.; Reischauer, S.; Chauhan, V.S.; Borkovich, M.; et al. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat. Commun. 2016, 7, 11303.
- Ghazizadeh, Z.; Kiviniemi, T.; Olafsson, S.; Plotnick, D.; Beerens, M.E.; Zhang, K.; Gillon, L.; Steinbaugh, M.J.; Barrera, V.; Sui, S.H.; et al. Metastable Atrial State Underlies the Primary Genetic Substrate for MYL4 Mutation-Associated Atrial Fibrillation. Circulation 2020, 141, 301–312.
- Rattka, M.; Westphal, S.; Gahr, B.M.; Just, S.; Rottbauer, W. Spen deficiency interferes with Connexin 43 expression and leads to heart failure in zebrafish. J. Mol. Cell. Cardiol. 2021, 155, 25–35.
- Chi, N.C.; Shaw, R.M.; Jungblut, B.; Huisken, J.; Ferrer, T.; Arnaout, R.; Scott, I.; Beis, D.; Xiao, T.; Baier, H.; et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008, 6, e109.
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008, 22, 734–739.
- Chi, N.C.; Bussen, M.; Brand-Arzamendi, K.; Ding, C.; Olgin, J.E.; Shaw, R.M.; Martin, G.R.; Stainier, D.Y. Cardiac conduction is required to preserve cardiac chamber morphology. Proc. Natl. Acad. Sci. USA 2010, 107, 14662–14667.
- Jou, C.J.; Arrington, C.B.; Barnett, S.; Shen, J.; Cho, S.; Sheng, X.; McCullagh, P.C.; Bowles, N.E.; Pribble, C.M.; Saarel, E.V.; et al. A Functional Assay for Sick Sinus Syndrome Genetic Variants. Cell. Physiol. Biochem. 2017, 42, 2021–2029.
- De Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.F.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641.
- Langheinrich, U.; Vacun, G.; Wagner, T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol. Appl. Pharmacol. 2003, 193, 370–382.
- Hassel, D.; Scholz, E.P.; Trano, N.; Friedrich, O.; Just, S.; Meder, B.; Weiss, D.L.; Zitron, E.; Marquart, S.; Vogel, B.; et al. Deficient zebrafish ether-à-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation 2008, 117, 866–875.
- Pineda, S.; Nikolova-Krstevski, V.; Leimena, C.; Atkinson, A.J.; Altekoester, A.K.; Cox, C.D.; Jacoby, A.; Huttner, I.G.; Ju, Y.K.; Soka, M.; et al. Conserved Role of the Large Conductance Calcium-Activated Potassium Channel, K(Ca)1.1, in Sinus Node Function and Arrhythmia Risk. Circ. Genom. Precis Med. 2021, 14, e003144.
- Langenbacher, A.D.; Shimizu, H.; Hsu, W.; Zhao, Y.; Borges, A.; Koehler, C.; Chen, J.N. Mitochondrial Calcium Uniporter Deficiency in Zebrafish Causes Cardiomyopathy With Arrhythmia. Front. Physiol. 2020, 11, 617492.
- Harrington, J.K.; Sorabella, R.; Tercek, A.; Isler, J.R.; Targoff, K.L. Nkx2.5 is essential to establish normal heart rate variability in the zebrafish embryo. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R265–R271.
- Collins, M.M.; Ahlberg, G.; Hansen, C.V.; Guenther, S.; Marín-Juez, R.; Sokol, A.M.; El-Sammak, H.; Piesker, J.; Hellsten, Y.; Olesen, M.S.; et al. Early sarcomere and metabolic defects in a zebrafish pitx2c cardiac arrhythmia model. Proc. Natl. Acad. Sci. USA 2019, 116, 24115–24121.
- Kamel, S.M.; van Opbergen, C.J.M.; Koopman, C.D.; Verkerk, A.O.; Boukens, B.J.D.; de Jonge, B.; Onderwater, Y.L.; van Alebeek, E.; Chocron, S.; Polidoro Pontalti, C.; et al. Istaroxime treatment ameliorates calcium dysregulation in a zebrafish model of phospholamban R14del cardiomyopathy. Nat. Commun. 2021, 12, 7151.
- Tessadori, F.; Roessler, H.I.; Savelberg, S.M.C.; Chocron, S.; Kamel, S.M.; Duran, K.J.; van Haelst, M.M.; van Haaften, G.; Bakkers, J. Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis. Model Mech. 2018, 11, dmm035469.
- Huttner, I.G.; Trivedi, G.; Jacoby, A.; Mann, S.A.; Vandenberg, J.I.; Fatkin, D. A transgenic zebrafish model of a human cardiac sodium channel mutation exhibits bradycardia, conduction-system abnormalities and early death. J. Mol. Cell. Cardiol. 2013, 61, 123–132.
- Ebert, A.M.; Hume, G.L.; Warren, K.S.; Cook, N.P.; Burns, C.G.; Mohideen, M.A.; Siegal, G.; Yelon, D.; Fishman, M.C.; Garrity, D.M. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc. Natl. Acad. Sci. USA 2005, 102, 17705–17710.
- Garrity, D.M.; Childs, S.; Fishman, M.C. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development 2002, 129, 4635–4645.
- Koopman, C.D.; De Angelis, J.; Iyer, S.P.; Verkerk, A.O.; Da Silva, J.; Berecki, G.; Jeanes, A.; Baillie, G.J.; Paterson, S.; Uribe, V.; et al. The zebrafish grime mutant uncovers an evolutionarily conserved role for Tmem161b in the control of cardiac rhythm. Proc. Natl. Acad. Sci. USA 2021, 118, e2018220118.
- Ahlberg, G.; Refsgaard, L.; Lundegaard, P.R.; Andreasen, L.; Ranthe, M.F.; Linscheid, N.; Nielsen, J.B.; Melbye, M.; Haunso, S.; Sajadieh, A.; et al. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat. Commun. 2018, 9, 4316.
- Kleinhans, D.S.; Lecaudey, V. Standardized mounting method of (zebrafish) embryos using a 3D-printed stamp for high-content, semi-automated confocal imaging. BMC Biotechnol. 2019, 19, 68.
- Von der Heyde, B.; Emmanouilidou, A.; Mazzaferro, E.; Vicenzi, S.; Höijer, I.; Klingström, T.; Jumaa, S.; Dethlefsen, O.; Snieder, H.; de Geus, E.; et al. Translating GWAS-identified loci for cardiac rhythm and rate using an in vivo image- and CRISPR/Cas9-based approach. Sci. Rep. 2020, 10, 11831.
- Power, R.M.; Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 2017, 14, 360–373.
- Gunawan, F.; Gentile, A.; Gauvrit, S.; Stainier, D.Y.R.; Bensimon-Brito, A. Nfatc1 Promotes Interstitial Cell Formation During Cardiac Valve Development in Zebrafish. Circ. Res. 2020, 126, 968–984.
- Bensimon-Brito, A.; Boezio, G.L.M.; Cardeira-da-Silva, J.; Wietelmann, A.; Ramkumar, S.; Lundegaard, P.R.; Helker, C.S.M.; Ramadass, R.; Piesker, J.; Nauerth, A.; et al. Integration of multiple imaging platforms to uncover cardiovascular defects in adult zebrafish. Cardiovasc. Res. 2021, cvab310.
This entry is offline, you can click here to edit this entry!
