Atypical femoral fractures (AFF) are rare fragility fractures in the subtrocantheric or diaphysis femoral region associated with long-term bisphosphonate (BP) treatment. The etiology of AFF is still unclear even though a genetic basis is suggested.
- atypical femoral fractures
- bisphosphonates
1. Introduction
2. Variant Selection
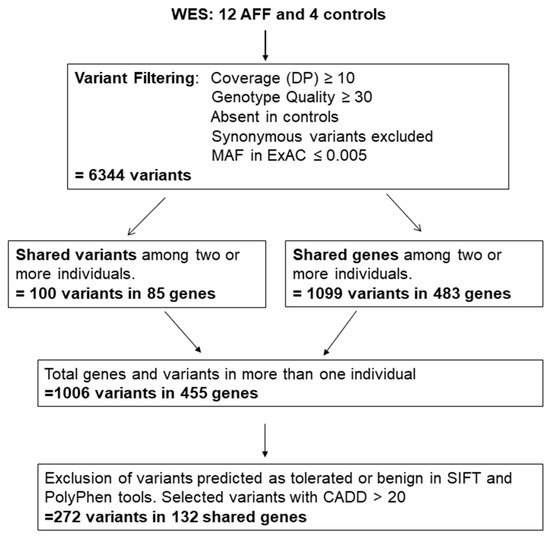
| Genes with Rare Variants in Two AFF Cases | Genes with Rare Variants in More Than two AFF Cases | ||||
|---|---|---|---|---|---|
| Two Different Variants | One Variant | ||||
| Gene Name | Gene Name | Gene Name | Gene Name | Gene Name | Number of Variants and (Carriers) |
| AASS | DNAH10 | PSD3 | ACADL | C8orf46 | 1 (3) |
| ABCA10 | DNAH12 | PTH1R | C1orf87 | CHRNG | 3 (3) |
| ABCA4 | DNAH6 | PYHIN1 | CD1A | DAAM2 | 3 (3, one homoz) |
| ABL2 | DYSF | R3HDML | CITED4 | DNAH14 | 4 (4) |
| ADAMTS12 | EFHB | RET | GBA | DNAH2 | 3 (3) |
| ANAPC11 | EP400 | RMDN1 | IQSEC3 | DNAH9 | 3 (3) |
| ANK3 | ERCC5 | RNF157 | NSMAF | FSIP2 | 3 (3) |
| ANKRD40 | FAT4 | RNF34 | PPP2R1B | HLA-DRB1 | 2 (4) |
| ARHGEF18 | FBLN7 | RTEL1 | SERPINB2 | HRASLS | 1 (3) |
| ARID1B | FLJ00418 | SCN9A | SPTBN1 | IGFLR1 | 2 (2, one homoz) |
| ASH1L | GBP3 | 5-Sep | SYDE1 | KRT10 | 1 (5) |
| ATAD2 | GPX4 | SH3BP2 | TNFRSF25 | LAMA1 | 3 (3) |
| ATP10B | HK3 | SHROOM4 | TRAPPC2L | LRP5 | 4 (3) |
| BIN1 | HPS6 | SIRT5 | TRIM32 | MRPS12 | 1 (3) |
| C10orf54 | IGFN1 | SLC26A9 | NEB | 4 (4) | |
| C12orf42 | IGSF10 | SLC2A7 | OBSCN | 5 (5) | |
| C14orf159 | IGSF22 | SLC34A3 | TCOF1 | 3 (4) | |
| C17orf107 | KLHL33 | SLC52A2 | TNXB | 3 (3) | |
| C6 | LLGL1 | SPTBN5 | TTN | 8 (8) | |
| C9orf84 | MEX3D | SRCAP | UTRN | 3 (3) | |
| CA9 | MKS1 | TAF15 | VEGFB | 1 (3) | |
| CDC42BPG | MMP20 | TENM4 | ZC3H3 | 3 (3) | |
| CERKL | MSLNL | TJP3 | |||
| CHAMP1 | NOD2 | TMEM143 | |||
| CLCN2 | NUP153 | TNRC6B | |||
| CRYBA1 | OPLAH | TOPORS | |||
| CTSE | PACSIN2 | TSFM | |||
| CUL7 | PARD6B | TTC14 | |||
| CYYR1 | PCDHAC1 | ZNF34 | |||
| DAB2IP | PDE4DIP | ZNF646 | |||
| DAW1 | PISD | ZNF729 | |||
| DHX34 | PLA2G4D | ZSCAN32 | |||
| Gene ID | Number of Carriers | Function | Bone Association | Bibliography Source |
|---|---|---|---|---|
| CUL7 | 2 | A core component of the 3 M complex required to regulate microtubule dynamics and genome integrity | Mutations in this gene produce the 3 m syndrome, which causes skeletal abnormalities | Genecards |
| DAAM2 | 3 | Involved in the canonical Wnt signaling, a pathway critical for bone formation and repair | SNPs in this gene are associated with estimated bone mineral density (eBMD). Daam2 knockout mouse showed decreased bone strength |
Musculoskeletal Knowledge Portal, Morris et al., 2019 [24] |
| DNAH10 | 2 | Found in cilia and flagella; ATPase activity and microtubule motor activity | SNPs in this gene are associated with waist-hip ratio and eBMD. | Musculoskeletal Knowledge Portal |
| DNAH12 | 2 | ATPase activity and microtubule motor activity | SNPs in this gene are associated with waist-hip ratio and eBMD | Musculoskeletal Knowledge Portal |
| LAMA1 | 3 | A major component of the basal membrane which has been implicated in a wide variety of biological processes including cell adhesion, differentiation, migration, and signaling | Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components. | Genecards |
| LRP5 | 4 | A co-receptor with Frizzled protein family members for transducing signals by Wnt proteins | It plays a key role in skeletal homeostasis and many bone density related diseases are caused by mutations in this gene | Genecards |
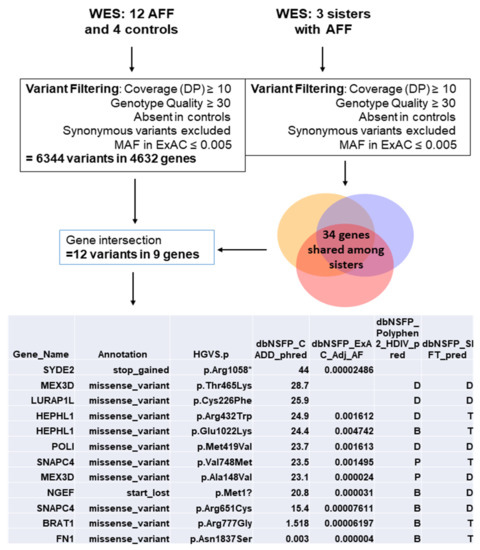
| MEX3D | 2 | RNA binding protein, may be involved in post-transcriptional regulatory mechanisms | Found mutated in three sisters with AFF | Roca-Ayats N, et al. 2018 [12] |
| PTH1R | 2 | A receptor for parathyroid hormone (PTH) and for parathyroid hormone-like hormone (PTHLH). | Involved in the Hedgehog and PTH signaling pathways in bone and cartilage development | Genecards |
| SLC34A3 | 2 | Involved in the transporting phosphate into cells via sodium cotransport in the renal brush border membrane, and contributes to the maintenance of inorganic phosphate concentration in the kidney | Mutations in this gene are associated with hereditary hypophosphatemic rickets with hypercalciuria. | Genecards |
| SPTBN1 | 2 | Spectrin is an actin crosslinking and molecular scaffold protein that links the plasma membrane to the actin cytoskeleton, and functions in the determination of cell shape, arrangement of transmembrane proteins, and organization of organelles | SNPs in this gene are associated with eBMD and total body BMD | Musculoskeletal Knowledge Portal |
| TNRC6B | 2 | Involved in cellular senescence, innate or adaptive immune system, Wnt signaling, and calcium modulating pathways | SNPs in this gene are mainly associated with lean mass. One SNP was also associated with lower lumbar spine BMD and increased risk of fractures | Karasik D, et al. 2019 [27] |
| TNXB | 3 | A member of the tenascin family of extracellular matrix glycoproteins | Mutations in this gene are associated with the Ehlers-Danlos Syndrome | Genecards |
3. AFF Network Analysis with Candidate Genes
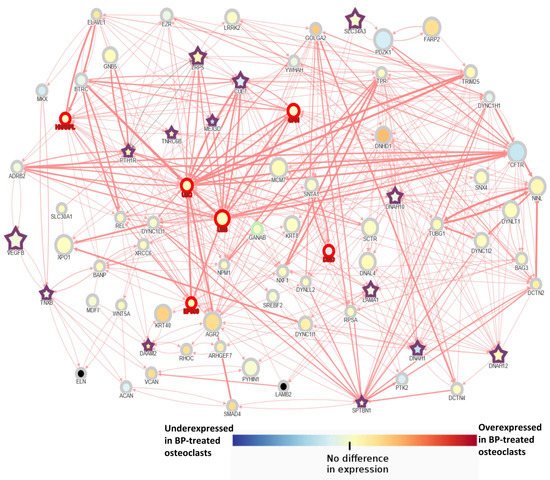
The main conclusion of this study is that AFF may present a multigenic background, specific to each patient, in which an accumulation of susceptibility variants may lead to a predisposition to BP-related AFF. Our analysis suggested that Wnt signaling may play a relevant role in the BP-related AFFs as half of the patients had mutations in a gene of this pathway. In silico analysis suggested a complex interaction network among the different mutated genes as well as a biological enrichment for cytoskeleton and cilium organization. WES analysis provided evidence to support the hypothesis that several genes and their interactions may be involved in the development of AFF, and, along with BP treatment and, in some cases, glucocorticoids, they may trigger the perfect storm.
This entry is adapted from the peer-reviewed paper 10.3390/genes13010146
References
- John A. Morris; 23andMe Research Team; John P. Kemp; Scott E. Youlten; Laetitia Laurent; John G. Logan; Ryan C. Chai; Nicholas A. Vulpescu; Vincenzo Forgetta; Aaron Kleinman; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nature Genetics 2018, 51, 258-266, 10.1038/s41588-018-0302-x.
- Hanh H Nguyen; Denise M Van De Laarschot; Annemieke Jmh Verkerk; Frances Milat; M Carola Zillikens; Peter R Ebeling; Genetic Risk Factors for Atypical Femoral Fractures (AFFs): A Systematic Review. JBMR Plus 2018, 2, 1-11, 10.1002/jbm4.10024.
- Fjorda Koromani; Katerina Trajanoska; Fernando Rivadeneira; Ling Oei; Recent Advances in the Genetics of Fractures in Osteoporosis. Frontiers in Endocrinology 2019, 10, 337, 10.3389/fendo.2019.00337.
- Neus Roca-Ayats; Pei Ying Ng; Natàlia Garcia-Giralt; Maite Falcó-Mascaró; Mónica Cozar; Josep Francesc Abril; José Manuel Quesada Gómez; Daniel Prieto-Alhambra; Xavier Nogués; James E Dunford; et al. Functional Characterization of a GGPPS Variant Identified in Atypical Femoral Fracture Patients and Delineation of the Role of GGPPS in Bone-Relevant Cell Types. Journal of Bone and Mineral Research 2018, 33, 2091-2098, 10.1002/jbmr.3580.
- Isabel Pérez-Núñez; José L. Pérez-Castrillón; María T. Zarrabeitia; Carmen Garcia-Ibarbia; Laura Martínez-Calvo; José M. Olmos; Laisa S. Briongos; Javier Riancho; Victoria Camarero; Josep M. Muñoz Vives; et al. Exon array analysis reveals genetic heterogeneity in atypical femoral fractures. A pilot study. Molecular and Cellular Biochemistry 2015, 409, 45-50, 10.1007/s11010-015-2510-3.
- Francesca Marini; Alberto Falchetti; Sandra Silvestri; Yu Bagger; Ettore Luzi; Annalisa Tanini; Claus Christiansen; Maria Luisa Brandi; Modulatory effect of farnesyl pyrophosphate synthase (FDPS) rs2297480 polymorphism on the response to long-term amino-bisphosphonate treatment in postmenopausal osteoporosis. Current Medical Research and Opinion 2008, 24, 2609-2615, 10.1185/03007990802352894.
- Hyung Jin Choi; Ji Yeob Choi; Sun Wook Cho; Daehee Kang; Ki Ok Han; Sang Wan Kim; Seong Yeon Kim; Yoon-Sok Chung; Chan Soo Shin; Genetic Polymorphism of Geranylgeranyl Diphosphate Synthase (GGSP1) Predicts Bone Density Response to Bisphosphonate Therapy in Korean Women. Yonsei Medical Journal 2010, 51, 231-8, 10.3349/ymj.2010.51.2.231.
- José M. Olmos; Maria T Zarrabeitia; Jose Luis Hernandez; Carolina Sanudo; Jesus Gonzalezmacias; Jose A Riancho; Common allelic variants of the farnesyl diphosphate synthase gene influence the response of osteoporotic women to bisphosphonates. The Pharmacogenomics Journal 2010, 12, 227-232, 10.1038/tpj.2010.88.
- Mohammad Kharazmi; Karl Michaëlsson; Jörg Schilcher; Niclas Eriksson; Håkan Melhus; Mia Wadelius; Pär Hallberg; A Genome-Wide Association Study of Bisphosphonate-Associated Atypical Femoral Fracture. Calcified Tissue International 2019, 105, 51-67, 10.1007/s00223-019-00546-9.
- Francesca Marini; Maria Luisa Brandi; Atypical femur fractures: a distinctive tract of adult hypophosphatasia. Clinical Cases in Mineral and Bone Metabolism 2017, 14, 324-328, 10.11138/ccmbm/2017.14.3.324.
- Symeon Tournis; Anastasia D. Dede; Osteogenesis imperfecta – A clinical update. Metabolism 2017, 80, 27-37, 10.1016/j.metabol.2017.06.001.
- Neus Roca-Ayats; Pei Ying Ng; Natàlia Garcia-Giralt; Maite Falcó-Mascaró; Mónica Cozar; Josep Francesc Abril; José Manuel Quesada Gómez; Daniel Prieto-Alhambra; Xavier Nogués; James E Dunford; et al. Functional Characterization of a GGPPS Variant Identified in Atypical Femoral Fracture Patients and Delineation of the Role of GGPPS in Bone-Relevant Cell Types. Journal of Bone and Mineral Research 2018, 33, 2091-2098, 10.1002/jbmr.3580.
- Isabel Pérez-Núñez; José L. Pérez-Castrillón; María T. Zarrabeitia; Carmen Garcia-Ibarbia; Laura Martínez-Calvo; José M. Olmos; Laisa S. Briongos; Javier Riancho; Victoria Camarero; Josep M. Muñoz Vives; et al. Exon array analysis reveals genetic heterogeneity in atypical femoral fractures. A pilot study. Molecular and Cellular Biochemistry 2015, 409, 45-50, 10.1007/s11010-015-2510-3.
- John A. Morris; 23andMe Research Team; John P. Kemp; Scott E. Youlten; Laetitia Laurent; John G. Logan; Ryan C. Chai; Nicholas A. Vulpescu; Vincenzo Forgetta; Aaron Kleinman; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nature Genetics 2018, 51, 258-266, 10.1038/s41588-018-0302-x.
- Hanh H Nguyen; Denise M Van De Laarschot; Annemieke Jmh Verkerk; Frances Milat; M Carola Zillikens; Peter R Ebeling; Genetic Risk Factors for Atypical Femoral Fractures (AFFs): A Systematic Review. JBMR Plus 2018, 2, 1-11, 10.1002/jbm4.10024.
- Fjorda Koromani; Katerina Trajanoska; Fernando Rivadeneira; Ling Oei; Recent Advances in the Genetics of Fractures in Osteoporosis. Frontiers in Endocrinology 2019, 10, 337, 10.3389/fendo.2019.00337.
